Physics-Biology interface seminar – archives
Breaking barriers: nanomedicine fate from administration target site
S. Mura, Orsay/France
Nanoscale systems for drug delivery have the potential to overcome the limitations of conventional treatments, thus providing a solution to unmet medical needs.
The therapeutic benefits of this approach have led to the commercialization of more than 50 nanomedicines such as doxorubicin-loaded liposomes (Doxil®, Myocet®), paclitaxel-albumin nanoparticles (Abraxane®), and more recently, lipid carriers for the delivery of siRNA (Patisiran/ONPATTRO®) or mRNA (BioNTech/Pfizer and Moderna COVID-19 vaccines).
Although these results provide clear evidence of the potential of nanomedicines for the efficient delivery of chemotherapeutics, there is still a substantial gap between the favorable preclinical results and the real clinical performances.
The introduction of nanomedicine in the clinic has been partly hampered by the lack of effective delivery to the target in vivo. Among the multiplicity of imputable factors, a major role can be attributed to: (i) the modifications undergone by nanomedicines after interaction with molecules/proteins in the bloodstream that endow them with a specific molecular signature and (ii) the numerous biological barriers that these nanomedicines must cross (e.g., the vascular endothelium, the tumor extracellular matrix, etc…).
In this context, it is necessary to have a clearer comprehension of their fate after administration.
Our research group is focusing on this topic, and we are developing different tools to investigate and better understand the barriers encountered nanomedicines both in the circulation after intravenous administration and, after extravasation, in the complex tumor microenvironment.
During this talk, the most significant results we have obtained will be presented
Literature:
[1] Sobot D, Mura S, Yesylevskyy SO, Dalbin L, Cayre F, Bort G, Mougin J, Desmaële D, Lepetre-Mouelhi S, Pieters G, Andreiuk B, Klymchenko AS, Paul JL, Ramseyer C, Couvreur P. Nature Comm 2017, 8, 15678
[2] Lazzari G, Nicolas V, Matsusaki M, Akashi M, Couvreur P, Mura S. Acta Biomaterialia 2018, 78, 296.
[3] Bidan N, Dunsmore G, Ugrinic M, Bied M, Moreira M, Deloménie C, Ginhoux F, Blériot C, de la Fuente M, Mura S. Drug Deliv Transl Res. 2023
Migration and Seach of Immune Cells
Franziska Lautenschläger (Saarland University, Germany)
Beware the unusual seminar time
Immune cells are often called the ‚sentinels‘ of the immune system, constantly searching for any kind of pathogen. During this search, immune cells spread over a given area with the task of finding objects in the most efficient way. Parameters that influence this search are migration speed, migration persistence and the correlation between the two [1]. These parameters can be modulated to diversify search strategies [2]. We have investigated how we can modify either the cellular environment or the cellular cytoskeleton (all types) to control migration and the search efficiency [3, 4].
- Shaebani, M.R., et al., Persistence-Speed Coupling Enhances the Search Efficiency of Migrating Immune Cells. Phys Rev Lett, 2020. 125(26): p. 268102.
- Shaebani, M.R., M. Piel, and F. Lautenschläger, Distinct speed and direction memories of migrating dendritic cells diversify their search strategies. Biophys. J., 2022. 121(21): p. 4099-4108.
- Shaebani, M.R., et al., Effects of vimentin on the migration, search efficiency, and mechanical resilience of dendritic cells. Biophys. J., 2022. 121(20): p. 3950-3961.
- Sadjadi, Z., et al., Ameboid cell migration through regular arrays of micropillars under confinement. Biophys J, 2022. 121(23): p. 4615-4623.
Biological tissues as materials: fluidity, rigidity and rheology
Cristina Marchetti (UCSB, USA)
Joint seminar with Institut Curie Orsay. Note the unusual location and time.Over the last decade there has been growing evidence that dense biological tissue can spontaneously undergo transitions between a solid-like rigid state and a fluid-like state. The rheological state of the tissue in turn influences the transmission of mechanical deformations, which plays a central role in driving developmental processes, such as wound healing and morphogenesis, and tumor progression. In this talk I will describe our computational and analytical investigations of the linear and nonlinear mechanical response of confluent tissue to shear deformations.
Semantic self-assembly in a meaning-less energy landscape far from equilibrium (seminar on Zoom)
Pablo Sartori (Gulbenkian Institute for Molecular Medicine)
Due to an illness from the speaker, the seminar has been converted to a remote format - see Zoom link below. Some of us will gather in the seminar room to watch. Feel free to join us.Like the letters in the alphabet forming words, reusing components is an efficient strategy for assembling a large number of target structures from heterogeneous mixture of components. Examples range from synthetic DNA origami to proteins self-assembling into complexes. The standard self-assembly paradigm views target structures as free-energy minima of a mixture. While this is an appealing picture, at high speed structures may be kinetically trapped in local minima, reducing self-assembly accuracy. How then can high speed, high accuracy, and combinatorial usage of components coexist? We propose to reconcile these three concepts not by avoiding kinetic traps, but by exploiting them to encode target structures. This can be achieved by sculpting the kinetic pathways of the mixture, instead of its free-energy landscape. In other words, targets structures with semantic value are not enconded in the energy landscape, but rather on the kinetics. We formalize these ideas in a minimal toy model, for which we analytically estimate the encoding capacity and kinetic characteristics, in agreement with simulations. Our results may be generalized to other soft-matter systems capable of computation, such as liquid mixtures or elastic networks, and pave the way for high-dimensional information processing far from equilibrium.
Time: May 16, 2025 11:00 AM Paris https://cnrs.zoom.us/j/93701191981?pwd=abTkbFq72axppbR2pHUgIumGG6Ojk0.1 Meeting ID: 937 0119 1981 Passcode: Ufwf5x
Labeling nanoparticles with contrast agents to trace their accumulation to diseased tissues
Nicolas Tsapis (Institut Galien)
In drug delivery, nanomedicines are often associated with a contrast agent to yield nanotheranostic systems allowing both imaging and treatment. The theranostic word was first mentioned in 2002 and comes from the contraction of therapy and diagnostics. Nanotheranostics are designed to image nanoparticle biodistribution, to survey the extent of the disease, to deliver the treatment and to monitor in real time its mechanism of action and its efficacy. In this presentation, we will focus on the first goal and will present several examples of labeling polymer nanoparticles to detect them with one of three different imaging modalities: ultrasound, 19F magnetic resonance imaging or photoacoustics.
SEMINAR CANCELLED
Franziska Lautenschläger (U. Saarlandes, Germany)
Establishment of phyllotaxy from a single cell in the moss Physcomitrium Patens
Laure Mancini (ENS Lyon)
Talk by the winner of the PhysBio2024 best junior talk competition. The speaker will join us over Zoom, but those interested can join us in the LPTMS seminar room to watch it. The shape of living organisms follows robust architectural plans that determine the main axis and the relative position of each organ. In plants, organs such as leaves arrange themselves around the stem with a mathematical regularity, forming large-scale patterns like spirals. This organization is called phyllotaxis and emerges thanks to the activity of stem cells located at the apical meristem, which corresponds to the tip of the leafy shoot. Depending on the group of plant, the apical meristem can be multi- or unicellular. In the case of the moss Physcomitrium Patens, this structure is composed of a single stem cell, the apical cell (AC), which is responsible for producing all the leaves. As a result, the position of each new leaf strongly depends on the division pattern of the AC. However, the mechanisms that control the positioning of the apical cell division plane to pattern moss phyllotaxis remain to be discovered. As an entry point to the problem, we studied the role of auxin signaling in this context because, the hormone auxin is known to be instrumental in establishing phyllotactic patterns in multicellular meristems. We asked if this hormone can also act at the scale of a unique stem cell, to insure a robust phyllotactic pattern. To do so, we used a combination of quantitative time-lapse imaging, genetic tools and mutants as well as cellular shape analysis and modeling. With these approaches, we found that auxin is involved in the robustness of the spiral phyllotaxy of the moss and that it affects cellular properties such as growth anisotropy, that can impact the global architecture of the moss leafy shoot(Non-)Reciprocal interactions in oriented mixtures
Joseph Ackermann (LJP, Sorbonne U.)
Joint seminar with LPS - note the unusual location and its change compared with previous announcementsInteractions between the components determine the properties of mixtures. In particular, non-reciprocal phase transitions have attracted attention in recent years due to features such as time-dependent phases [1]. In the context of cancer, the heterogeneity of the environment requires the consideration of the interplay between different components. In particular, polar and nematic orientations are ubiquitous. In this talk, I will present two experiments and their physical description involving reciprocal and non-reciprocal nematic or polar interactions and some questions they raise [2,3].
[1] Fruchart, Michel, et al. "Non-reciprocal phase transitions." Nature 592.7854 (2021): 363-369.
[2] Jacques, Cécile, et al. "Aging and freezing of active nematic dynamics of cancer-associated fibroblasts by fibronectin matrix remodeling." bioRxiv (2023): 2023-11.
[3] Bell, Samuel, et al. "Ordering, spontaneous flows and aging in active fluids depositing tracks." arXiv preprint arXiv:2409.05195 (2024).
Electrostatic zipper in curved DNA condensates
Amélie Leforestier (U. Paris-Saclay)
Joint seminar with LPS: beware of the unusual locationIn helical polyelectrolytes like DNA, the electrostatic zipper is a model of interaction, which, taking into account the helical surface charge distribution, predicts mutual azimuthal orientation of close-packed molecules. This phenomenon is poorly documented experimentally. In particular, electrostatic zipper is not compatible with curvature, a general feature of dense states of DNA, both in vitro and in vivo. Using cryo-EM imaging of DNA toroids, we analyze the relationship between curvature, helical correlations, and DNA helical pitch. We show that in phase helical alignments are preferred over a wide range of ionic concentrations. Optimization of this preferred configuration is associated to rearrangements within the toroid as it grows, with establishment of the EZ followed by a global polygonal shaping. In addition, a local decrease in the DNA helical pitch is measured in highest curvature regions. This phenomenon, at work in DNA toroids, is also likely to occur in spheroidal globules, bacteriophage chromosomes, and, more generally, all forms of curved condensed DNA – and other helical polyelectrolytes – both in vivo and in vitro. In particular, it should be taken into account in material design involving curved DNA origami.
Ajay Subbaroyan (IMSc Chennai, India)
Uncovering and leveraging design principles in Boolean models of gene regulatory networks
Beware of the unusual day and timeslot Boolean networks (BNs) are a well-established framework for modeling the dynamics of gene regulatory networks (GRNs). In Boolean GRNs, genes can be either "on" or "off", with their dynamics dictated by regulatory logic rules or Boolean functions (BFs). The complete dynamics of BNs are encapsulated in its state transition graph (STG). In this presentation, I will aim to address three research lacunae. The first is whether regulatory logic rules in reconstructed BNs that model cellular decision making, are random. We show that certain biologically meaningful BFs, namely, the read-once functions (RoFs) and the nested canalyzing functions (NCFs) are highly preponderant in such reconstructed BNs. This observation is substantiated by proving that RoFs and NCFs possess minimal complexity based on two metrics, Boolean complexity and average sensitivity, respectively. The second is the proposal of a model selection framework for developmental Boolean GRNs. Our framework leverages relative stability constraints that are predicated on the hierarchy of cell states on the developmental landscape. The application of this framework to the latest reconstructed Boolean GRN of the Arabidopsis thaliana root stem cell niche yields several improved models. The third is how various types of regulatory logic rules influence the structure of the STG of a BN. Certain biologically meaningful BFs typically engender more "bushy" and convergent STGs, indicative of robust dynamics, as opposed to STGs obtained using random BFs. This investigation is made possible by our adaptation of measures from the theory of cellular automata to the domain of BNs.Protein-membrane interactions with a twist
Martin Michael Müller (U. de Lorraine)
To understand how a biofilament can interact with a lipid membrane, mesoscopic models are of particular interest. Within a framework of elasticity theory and geometry, two mechanisms have been proposed, which can induce membrane deformations due to a biofilament: the Twister and the Darboux torque mechanism [1]. Whereas the Darboux torque mechanism has been shown to explain membrane deformations by a polymer in several important biological systems, the Twister mechanism has been studied more carefully only recently [2].
In my talk I will discuss how the Twister mechanism together with numerical simulations can explain the membrane translocation of botulinium toxins, which are among the most powerful toxins produced in nature. The initial deformation of the membrane by the toxin is caused by the presence of local torques arising from asymmetric positions of hydrophobic residues. Different torque distributions are observed in the simulations and permit an origin for the mechanism opening the membrane to be proposed.
[1] J. Fierling et al. Soft Matter, 12:5747, 2016.
[2] A. Delort et al. IJMS, 25:2481, 2024.
Catching up: towards bio-inspired catch-bonded materials
Jasper van der Gucht (Wageningen U., the Netherlands)
Many biological materials face conflicting mechanical demands: they must be dynamic and adaptable on the one hand, and robust and resilient on the other. Dynamicity asks for labile bonds that readily dissociate and reform, while resilience asks for strong and long-lived bonds. Nature has found a solution for these seemingly contrasting requirements: catch bonds. While conventional bonds weaken under tension, catch bonds do the opposite: they strengthen under stress. Under quiescent conditions the bonds are weak, allowing for the dynamic rearrangements necessary for cell functioning, but under conditions of high stress they stabilize, thereby protecting the cells against mechanical stress. Since their discovery two decades ago, catch bonds are now recognized as a crucial design element for controlling cellular mechanics. Yet, how catch bonds exactly perform their task at the scale of networks and cells is not well understood.
In this presentation, I will discuss two recent approaches from our group that aim to shed insight on this. In the first part, I will discuss simulation results of catch-bonded polymer networks that show how catch bonds collectively protect networks against mechanical damage, and in particular how the mobility of the bonds influences this. In the second part, I will share our efforts to design a synthetic DNA-based catch bond, by using a combination of computational design, experiments and simulations.
- J. Ruiz-Franco, J. Tauber & J. van der Gucht, “Cross-linker Mobility Governs Fracture Behavior of Catch-Bonded Networks”, Physical Review Letters 130 (118203), 2023.
- M. van Galen, A. Bok, T. Peshkovsky, J. van der Gucht, B. Albada & J. Sprakel, “De novo DNA-based catch bonds”, Nature Chemistry, in press, 2024.
The evolution, structure and function of ESCRT-III
Buzz Baum (Cambridge University)
In this talk we will explore the evolution of the eukaryotic cytoskeleton. As we will see, much of the machinery, including actin, tubulin and ESCRT-III, has its origins in archaea. We can therefore use archaea as a simple model in which to understand the evolution, structure and function of ESCRT-III polymers in membrane remodelling and cytokinesis.Exploring Membrane Topology Transformations using Polymer Field Theory
Russell Spencer (University of Göttingen)
Membrane remodeling, including fusion and fission, plays a crucial role in various cellular processes. However, understanding the intricacies of these topological changes can be challenging due to the involvement of large-scale membrane rearrangements and their sensitivity to small-scale molecular behavior. Moreover, these remodeling events face significant free-energy barriers that necessitate the presence of catalytic proteins. In this work, we utilize self-consistent field theory (SCFT) in combination with the string method to identify the Minimum Free Energy Path (MFEP). We thereby determine the most probable pathway for specific remodeling transitions implicated in cellular signaling and organelle division. This approach has allowed us to discover a new pathway by which the fusion of membranes may catalyze their fission. Furthermore, we extend conventional SCFT methods, introducing proteins inspired by the dynamin family. These proteins facilitate fission by constricting membrane tubes. We find that the free energy barrier to fission depends strongly on membrane tension and constriction. In addition to simply constricting the membrane, dynamin's PH domains are inserted between lipid head groups, inducing membrane distortion. Our results emphasize the crucial role of this distortion in reducing the free energy barrier to fission. This research sheds light on the underlying mechanisms of membrane remodeling and provide insights into cellular processes involving topological changes.
How to create form and dynamics in a living embryo
Emily Gehrels (Aix-Marseille Université)
Beware of the unusual location
Biological organisms are able to develop from simple eggs to complex adult forms via a series of dynamic rearrangements, during which embryonic tissues generate stress, deform, and flow. What can we learn about the rules and possibilities of autonomous structure formation by studying their development? I will present one such study where we uncover how Drosophila embryos use symmetric forces to create a polarized flow of cells needed to achieve their final form.
Cell flows in the early Drosophila embryo are driven by an interplay between biological signaling and tissue mechanics. Using live imaging, we observe how changes in the presence of force-generating proteins and the geometry of the tissue relate to tissue dynamics at the onset of morphogenesis. We use theoretical and computational methods to model the behavior of the tissue and experimentally challenge our findings using select genetic perturbations of the embryos. With this combination of experimental and modeling approaches, we have uncovered how organized multicellular dynamics emerge from genetic, mechanical, and geometric “information” during early Drosophila development.
Simple Chromosome Partitioning Mechanisms and Dynamics of a Mitotic Spindle.
(Beware of the unusual location)
François Nédélec (Cambridge University)
We are using theory to explore simple mechanisms of chromosome partitioning with the aim of understanding the design principles of mitotic spindle assembly. Firstly, I will present how artificial evolution in a computer can uncover simple combinations of cytoskeletal elements that will self-organize to pull on a kinetochore pair symmetrically and reliably. I will then discuss the requirements to extend such elementary mechanisms to build a mitotic spindle that can handle multiple chromosomes. Secondly, I will present the characteristics of the mitotic spindles found in the roots of Arabidopsis thaliana, and argue that given these quantities, this plant spindle stands out as an ideal subject to build a quantitatively accurate 3D simulation. I will present our attempts to build such a model, detailing some key assumptions and using a simulation to illustrate the remarkable dynamics of a metaphase mitotic spindle.
Fast deformation under flow and morphological recovery of cancer cells
Catherine Villard (U. Paris-Cité)
Warning: joint seminar with LPS, takes place at LPS
The cancer metastatic cascade includes a blood circulation step for cells detached from the primary tumor. This stage involves significant shear stress as well as large and fast deformations as the cells circulate through the microvasculature. The recovery dynamics after deformation is pivotal to understand how a cell can pass through the multiple capillary constrictions encountered during a single hemodynamic cycle. I will present results on the recovery dynamics of circulating cells representative of different cancer phenotypes and their nuclei, obtained thanks to a dedicated microfluidic system we have recently developed. In brief, all cell types display an elastic recovery behavior followed by a slower viscoelastic regime, while nuclei, depending on the cell type, can strikingly recover purely elastically over a time scale of 20 ms. The fast elastic cell recovery regime is under the control of cell contractility ensured by the integrity of the cell acto-myosin cortex while the presence of a vimentin cage around the nuclei confers elastic properties to this organelle. I will finally discuss how these behaviors can possibly impact cell transit and survival into the blood circulation.
From interfacial tubulation in Alcanivorax borkumensis biofilms to folding of membranes with nematic order.
Carles Blanch Mercader ( Physico-Chimie Curie )
Assemblies of anisotropic cells can exhibit long-range orientational order and topological defects, which often influence processes of shape formation. In the first part of this talk, I will introduce conditions for a morphological transition between a spherical and a dendritic phase in Alcanivorax borkumensis biofilm-covered oil microdroplets. Experimental observations demonstrate preferential interfacial tubulation at topological defects; bacteria assemblies on these tubes exhibit long-range nematic order. The second part of this talk focuses on the role of order-curvature couplings on equilibrium states of a membrane with nematic order that are generated by an integer topological defect. For varying elastic parameters and boundary anchoring conditions, the instability of a flat disc towards an out-of-plane deformed surface will be discussed. Finally, I will argue how these results can be relevant for biological systems and show how bacteria biofilms can utilize topological defects to facilitate membrane deformation and enhance oil access.
The first part is a joint work with M. Prasad, N. Obana, S.-Z. Lin, K. Sakai, J. Prost, N. Nomura, J.-F. Rupprecht, J. Fattaccioli, and A.S. Utada. The second part is a joint work with C. Thibault, Q. Chaboche, and D.J.G. Pearce.
Taming gas-converting metalloenzymes: (spectro)electrochemical insights
Joint seminar with the Soft Matter group at LPS
Alexandre Ciaccafava (Marseille Molecular Science Institute, France)
Metalloenzymes are at the heart of a wide range of bioenergetic processes essential to all living organisms. They are responsible for the exchange and conversion of energy by making or breaking chemical bonds with the concomitant exchange of electrons (e–) and protons (H+). They are extremely efficient biocatalysts that convert with high efficiency and affinity a multitude of substrates, from the simplest molecule such as hydrogen (H2) to much larger polymeric entities (cellulose), opening the way to high-impact biotechnological applications. Because of the biodiversity of the microorganisms from which they are derived (notably extremophiles adapted to various unusual biotopes), redox metalloenzymes can operate under a wide range of experimental conditions. Most redox enzymes contain non-noble metals (Ni, Fe, Cu, Mo,… ), assembled in a more or less complex way in the active center where their catalytic activity is located or in electronic relays that transfer electrons to other physiological partners, constituting a larger set of multi-partner electron transfer chains. As a result, redox metalloenzymes can advantageously replace chemical catalysts based on noble metals, whose scarcity generates a consequent cost, limiting their large-scale development. This set of remarkable properties shared by redox metalloenzymes has fuelled both the desire to control their reactivity and the attention of chemists to create bio-inspired catalysts. However, a thorough understanding of the functioning of redox metalloenzymes is a prerequisite for taming their exceptional properties and ultimately for their integration into biotechnological processes.
Macromolecular condensation buffers intracellular water potential
Emmanuel Derivery (Cambridge U.)
Optimum protein function and biochemical activity critically depends on water availability inside cells. Macromolecules restrict the movement of “structured” water molecules in their hydration layers, reducing the available “free” bulk solvent and therefore the total thermodynamic potential energy of water, or water potential. Within concentrated macromolecular solutions like the cytosol, we found that modest changes in temperature greatly impact the water potential. We predicted that lower temperatures would reduce the available “free” intracellular water in a similar manner to external hyperosmotic conditions, and vice versa for higher temperatures and hypoosmotic conditions. We validated this duality of temperature and osmotic strength on cellular physiology: hypoosmotic conditions mimicked high temperature in thermosensitive yeast mutants, whereas cold temperatures induced chondrocyte Ca2+ signalling similar to hyperosmotic conditions. Most remarkably, simple manipulations of solvent thermodynamics were sufficient to prevent cell death upon extreme cold or heat shock. Physiologically, cells must sustain their activity in the face of fluctuating temperature, pressure and osmotic strength that impact water potential within seconds, but established mechanisms of water homeostasis act over much slower timescales, so we postulated the existence of a rapid compensatory response. We find this function is performed by water potential-driven changes in macromolecular assembly, particularly biomolecular condensation of intrinsically-disordered proteins, which is determined by the water potential rather than the concentration of any specific macromolecule. In cells, formation or dissolution of biomolecular condensates counteracted thermal or osmotic perturbations of water potential, which was robustly buffered in isolated cytoplasm. Our results indicate that biomolecular condensation constitutes an intrinsic biophysical feedback response that rapidly compensates for intracellular osmotic and thermal fluctuations. We suggest preserving water availability within the concentrated cytosol is an overlooked evolutionary driver of protein (dis)order and function.
The architecture and mechanism of lipid transfer at interfaces between lipid droplets
Iva Ganeva (University of Bern, Switzerland)
Lipid droplets (LDs) are intracellular organelles responsible for regulated storage and release of neutral lipids. LDs feature a neutral lipid core enclosed by a phospholipid monolayer and surface proteins involved in lipid metabolism. The protein Cidec is crucial for LD enlargement and accumulates at interfaces between LDs where it facilitates directional neutral lipid transfer from the smaller to the larger LD. Deciphering the structure of the LD-LD interface is key in understanding the molecular mechanism of lipid transfer. We use live cell imaging and electron cryo-tomography of human cells to visualize interfaces between LDs and understand the mechanism of lipid transfer directly inside cells. Furthermore, by biochemical reconstitutions using recombinant Cidec and various artificial membrane systems, we aim at uncovering the minimal machinery for interface architecture and lipid transfer function. Our current model is that lipid transfer occurs through largely intact monolayers at an interface which resembles an organelle contact site. We hypothesise that this process is facilitated by the ability of Cidec to induce lipid packing defects into the monolayers, thereby making them permeable for neutral lipids.
Statistical physics of molecular sorting in living cells
Joint seminar with the Soft Matter group at LPS
Elisa Floris (Politecnico de Torino, Italy)
Molecular sorting is a fundamental process responsible for the organization of intracellular matter: biomolecules that dwell on the outer and inner membranes of the cell are partitioned into spatially localized domains and engulfed into small vesicles, that detach from the membrane to be delivered to their right destinations. A recently proposed theoretical model of this process is based on the coupling of a) phase-separation-driven formation of sorting domains and b) domain-induced membrane bending, leading to vesicle nucleation [1]. In this minimal model, molecules are randomly inserted on the lipid membrane, can diffuse laterally and aggregate into domains, and are extracted as part of a domain once it has reached a characteristic extraction size, representing the average size of mature vesicles. The predictions of the theory and numerical investigations of a latticegas realization of the model show the existence of an optimal region of parameter space where sorting is most efficient. In the experiments, detected domains are commonly classified into two groups: productive domains, if their growth eventually terminates in the nucleation of a vesicle, and unproductive domains which rapidly dissolve. To interpret these data, we use central notions of the classical theory of phase separation, such as the boundary tension and the critical domain size. Comparison with experimental results shows that the statistical properties of productive and unproductive domains inferred from experimental data are in agreement with those predicted from numerical simulations of the model, compatibly with the hypothesis that molecular sorting is driven by a phase separation process. The model is further extended to account for the simultaneous sorting of a multiplicity of molecular species, revealing that, when keeping fixed the total incoming molecular flux, the average molecule residence time increases with the heterogeneity of the molecular pool according to a simple scaling law. Molecular sorting is further investigated by means of a simplified one-dimensional lattice-gas model, where the membrane is allowed to undergo the topological transformations corresponding to fusion and detachment of molecule-loaded vesicles.
References
[1] Marco Zamparo, Donatella Valdembri, Guido Serini, Igor V Kolokolov, Vladimir V Lebedev, Luca Dall’Asta, and Andrea Gamba. Optimality in self-organized molecular sorting. Physical Review Letters, 126(8):088101, 2021.Quantifying Uncertainty in Symmetric Particle-Based Models for Statistical Inference
Antonin Della Noce (Institut Gustave Roussy)
Abstract: Particle-based or Individual-Based Models (IBMs), initially developed for kinetic gas theory, have found applications across various scientific disciplines, including computational biology, for the purpose of explaining emergent macroscopic phenomena from microscopic interactions. In many applications, some parameters of the system need to be inferred from observation data carrying very partial information on the underlying population / particle assembly This presentation is divided in two parts. The initial part addresses the issue of parameter inference in scenarios where observational data provide limited information about the underlying population or particle assembly. It will discuss the propagation of this partial system knowledge into uncertainties associated with parameter values. The subsequent part focuses on the evaluation of the consistency of mean-field approximations, specifically within the framework of a model representing plant populations in competition for light, which is partially observed.
Bio: Antonin Della Noce obtained his Ph.D. in Applied Mathematics from the Laboratory of Mathematics and Computer Science for Complex Systems (MICS) at Université Paris-Saclay. His doctoral research focused on population dynamical systems. Following his Ph.D., Antonin collaborated with Institut Gustave Roussy to conduct biostatistical research aimed at predicting breast cancer toxicities through the use of high-throughput proteomics. Additionally, he worked with Hôpital Bichat on developing screening strategies for sequencing patients suspected of having connective tissue disorders.
Collective effects in epithelial cell elimination
Romain Levayer, Department of Developmental and Stem Cell Biology, Institut Pasteur, Paris, France
While the signals regulating apoptosis during development are rather well known, what sets the precise spatio-temporal distribution of cell death and the adjustment of cell elimination to local perturbations is not well understood. Similarly, how cells eventually engage in apoptosis and how the removal of cells from epithelial layer is orchestrated through cell extrusion remained poorly understood, especially in vivo. In this seminar, I will illustrate the multi-layered regulation of cell death through three examples, the first one describing the impact of mechanical stress on cell elimination, the second one on local feedbacks that can fine tune the distribution of cell elimination in space and time revealing interesting self-organising properties, and the last one focusing on the engagement in apoptosis and the coordination of cell extrusion by caspases. Altogether, I will propose a roadmap to build a more predictive understanding of cell death in vivo that can directly impact tissue homeostasis, morphogenesis, growth and clonal selection.
AFM force spectroscopy studies unravel how functional nanodomains on pathogenic bacteria drive selective recognition by host immune cells.
Albertus Viljoen (Louvain Institute of Biomolecular Science and Technology, UC Louvain)
Adhesion to host cells and tissues is an important first step in infection employed by many bacterial pathogens. In mycobacteria, surface hydrophobic properties and specialized receptor-ligand bonds determine how these pathogens adhere to host immune cells. We recently used fast quantitative imaging (QI) atomic force microscopy (AFM) combined with hydrophobic tips to quantitatively map hydrophobic properties of mycobacterial pathogens, at high spatial resolution1. We discovered that abundant polar outer membrane lipids modulate the nanoscale distribution of hydrophobicity on the bacterial surface, which could be modulated by treatments with specific antibiotics. This led us to hypothesize that the presence of nanodomains enriched in specific surface molecules have a functional role in pathogen-host interactions. We have now found support for this hypothesis by studying recognition of pathogenic mycobacteria by the pathogen recognition receptor DC-SIGN 2. By probing live mycobacteria with AFM tips modified with single tetramers of the soluble extracellular domain of DC-SIGN we observed that the receptor forms complexes with ligands on pathogenic and nonpathogenic mycobacteria. Moreover, the complexes are mechanically weak, rupturing at ~30 pN under physiological force loading rates. However, by relying on the specific force-extension signatures generated we could map the locations of single DC-SIGN ligands at a resolution of ~20 nm, which revealed concentration of ligands into dense nanodomains on pathogenic mycobacteria exclusively. Upon bacteria-host cell contact, ligand nanodomains induce the recruitment and clustering of DC-SIGN, resulting in mechanically stable adhesion. This work highlights the key role of clustering of both ligands on pathogenic bacteria and DC-SIGN host receptors in pathogen recognition, a mechanism that might be widespread in host-pathogen interactions.
- Viljoen, A., Viela, F., Kremer, L. & Dufrêne, Y. F. Fast chemical force microscopy demonstrates that glycopeptidolipids define nanodomains of varying hydrophobicity on mycobacteria. Nanoscale Horiz. 5, 944–953 (2020).
- Viljoen, A. et al. Nanoscale clustering of mycobacterial ligands and DC-SIGN host receptors are key determinants for pathogen recognition. Sci. Adv. accepted, (2023).
A family of integrable Hamiltonian systems of classical particles with arbitrary two-body scattering shifts
Benjamin Doyon (King's College)
Some special systems of classical particles are integrable in the sense of Liouville: there are as many conserved quantities, including the Hamiltonian itself, as there are particles. When this happens, the system is typically not chaotic, and many of its properties can be evaluated exactly. It is interesting to find many-body systems that are integrable: interaction potentials that can written for any number of particles N, and such that the system is integrable for all N. When the interaction is local enough, such a many-body integrable system has the property of elastic and factorised scattering: in any scattering event, the sets of in- and out-momenta are the same. The trajectories are still shifted from linear trajectories due to the interaction, but these shifts are "factorised" into two-body shifts, as if the many-body scattering happened as a succession of separate two-body scattering events. Amongst the known locally interacting integrable models, only very specific functions of momenta do play the role of two-body shifts. I will overview a new family of many-body integrable systems for which the scattering two-body function can be taken as any (differentiable, positive) function. It can be seen as a certain flow on interactions, starting from free particles, and produced by perturbing by bilinear terms in conserved densities and currents. If time permits, I will explain how we evaluate the thermodynamics of this system, which gives the classical version of the so-called thermodynamic Bethe ansatz, and its hydrodynamics, which gives generalised hydrodynamics.
Instabilities and Geometry of Growing Tissues
Doron Grossman (College de France - for this work; LadHyX, Ecole polytechnique - current)
We present a covariant continuum formulation of a generalized two-dimensional vertexlike model of epithelial tissues which describes tissues with different underlying geometries, and allows for an analytical macroscopic description. Using a geometrical approach and out-of-equilibrium statistical mechanics, we calculate both mechanical and dynamical instabilities of a tissue, and their dependences on various variables, including activity, and cell-shape heterogeneity (disorder). We show how both plastic cellular rearrangements and the tissue elastic response depend on the existence of mechanical residual stresses at the cellular level. Even freely growing tissues may exhibit a growth instability depending on the intrinsic proliferation rate. Our main result is an explicit calculation of the cell pressure in a homeostatic state of a confined growing tissue. We show that the homeostatic pressure can be negative and depends on the existence of mechanical residual stresses. This geometric model allows us to sort out elastic and plastic effects in a growing, flowing, tissue.
Nanofluidics for the study of viral particle transport
Léa Chazot-Franguiadakis (ENS Lyon)
From a physical point of view, viruses are biological objects with remarkable mechanical and thermodynamic properties. Their detection and characterisation are currently major public health issues. In this context, the transport of viruses through nanopores is of interest both from a fundamental and a technological point of view, as a method to probe viral properties. We use a mimetic approach of a biological pore, the nuclear pore complex, which is taken by some viruses to access the nucleus. It has an exceptional selectivity due to the presence of a network of polymers forming a dynamic gel within its central channel. To better understand the characteristics of this transport we build a simplified and controlled environment based on functionalized nanoporous membranes. The transport of viral particles is studied using an optical detection method with adjustable specificity. Our device allows real-time detection of single viral particle transport at the level of a single pore. Our approach allows us to address technological issues, such as the quantification of virus concentration, for which we have developed a versatile method with a low detection limit. It also enables to address fundamental questions about viruspore interactions. In particular, we have highlighted an aggregation phenomenon related to the strong confinement of viruses under flow in the nanopore. The modelling of this phenomenon provides access to interaction parameters between the viruses and between the viruses and the pore. Merci de contacter eric.raspaud@universite-paris-saclay.fr et antoine.fruleux@universite-paris-saclay.fr si vous désirez rencontrer l’oratrice/l’orateur ou déjeuner avec nous.Odd Cosserat elasticity in active materials
Debarghya Banerjee (MPIPKS, Dresden)
Stress-strain constitutive relations in solids with an internal angular degree of freedom can be modelled using the Cosserat or micropolar elasticity. In the theory of emph{chiral active} Cosserat materials one would find an emph{odd} elastic term in addition to the usual Cosserat term in the constitutive relations. In this talk, I will develop a natural extension of Cosserat materials to include odd elasticity and discuss the phenomenological effects of these terms. I will discuss about static elastic properties of such a solid, where, we show that static response to vortical stresses lead to strains that depend on both the Cosserat and odd elasticity. We then compute the dispersion of linear solutions in these odd-Cosserat materials, where, we find the presence of emph{exceptional points} in the dispersion relation. Finally, we show the effect of Cosserat and odd elastic terms on the polarization of Rayleigh edge waves. Merci de contacter eric.raspaud@universite-paris-saclay.fr et antoine.fruleux@universite-paris-saclay.fr si vous désirez rencontrer l’oratrice/l'orateur ou déjeuner avec nous.Microfluidics and “Organs-on-chip”: is it possible to mimic blood vessels?
Anne-Marie Haghiri (C2N)
“Organs-on-a-chip” aim to capture key functions that are indispensable for the physiological functioning of a specific organ by mimicking the tissue elements that perform these functions in vivo. In this context, microfluidics offers the ability to reconstruct in vitro biological microenvironments with all the biophysical and biochemical expected parameters including cellular coatings. Along this talk, I will focus on microfluidic devices aiming to mimic blood microvessels based on two complementary approaches. The first approach concerns the microfluidic wearable artificial lung that recovers on chip the physiological function of breath. A microfluidic oxygenator mainly consists of three layers with a thin gas permeable membrane sandwiched between a blood microcapillaries network and a gas network. To meet clinical demands, such device must therefore meet certain requirements, which are 1/ support high blood flow without reducing the gas transfer efficiency, 2/ minimize pressure drop and shear stress in the blood module, 3/ enhance hemocompatibility by coatings. Our device has been designed at the 4-inch wafer scale with very dense curved blood and oxygen microcapillaries separated by a 15 µm-thick porous membrane [1]. Such design allows reducing both priming volume while maintaining efficient gas exchange. Biomimetic blood flow paths with low shear rate also promotes sustainable endothelialization since cells can be maintained viable for up to 2 weeks after initial seeding. The simplicity of connecting different units in the stacked architecture has also been demonstrated for 3- or 5-unit stacked devices that exhibit remarkable performance with low primary volume, high oxygen uptake and carbon dioxide release and high flow rate up to 80 ml/min [2]. The second more conventional approach consists to generate microvasculature directly inside the chip where a tumor model is growing. Here, we developed a simple method to fabricate high aspect ratio 3D microfluidic devices, as geometrically optimized microenvironment for long-term culture and vascularization of 3D tumor models (i.e., spheroids). The role of the height of the central chamber on the growth of a heterotypic pancreatic tumor spheroid, made of cancer cells and fibroblasts is under study. We recently demonstrated that a 500µm-thick microfluidic chamber promotes more gradual cell growth and migration, thanks to the availability of space for spheroid evolution. Finally, I will conclude on the importance of innovative design/architecture coupled with adapted microfabrication processes. References: [1] A-M. Haghiri-Gosnet, Lyas Djeghlaf, Julie Lachaux, Alisier Paris, Gilgueng Hwang, European Patent EP18306405.4 (29 Oct. 2018) "Microfluidic gas exchange devices and methods for making same" [2] Julie Lachaux, Gilgueng Hwang, Nassim Arouche, Sina Naserian, Abdelmounaim Harouri, V. Lotito, C. Casari, T. Lok, J B. Menager, J. Issard, J. Guihaire, C. V. Denis, P. J. Lenting, A. I. Barakat, G. Uzan, O. Mercier, Anne Marie Haghiri-Gosnet, Lab Chip, 2021, 21, 4791 DOI:10.1039/d1lc00356a Merci de contacter eric.raspaud@universite-paris-saclay.fr et antoine.fruleux@universite-paris-saclay.fr si vous désirez rencontrer l’oratrice/l'orateur ou déjeuner avec nous.Principles of Protein Assembly, From Molecules to Cells
Emmanuel Levy (Weizmann Institute of Science, Israel)
Life processes involve an intricate choreography between tens of millions of protein building blocks that form the infrastructure of cells. Recent technological advances have revealed the catalogs of proteins present in various cells and organisms. However, understanding how these protein building-blocks assemble and work together is an extraordinarily complex task. I will present our work aimed at addressing this challenge. First, I will describe how stickiness, defined as the chemical propensity of protein surfaces to bind to each other, is tuned across entire proteomes to minimize promiscuous interactions. Second, I will show that even single point-mutations changing the surface stickiness of homo-oligomeric proteins can trigger their infinite folded-state self-assembly. Finally, I will describe the design of synthetic protein condensates with tractable and tunable phase behavior; and work-in-progress, on how we are using these condensates to identify genes globally regulating macromolecular self-assembly in cells.
From stem cells heterogeneity to tissue morphogenesis during vertebrate embryo development.
Bertrand Benazeraf (U. Paul Sabatier, Toulouse)
Understanding the formation of embryonic shapes is one of the fundamental themes in developmental biology and a significant challenge to better understand congenital diseases. Posterior axis elongation is the morphogenetic event that produces the typical head-to-tail elongated body shape of vertebrate embryos. While the mechanisms of tissue elongation have mostly been studied in separate tissue types, the principles allowing for the coordination of elongation between tissues remain largely unknown. By using a combination of classical embryological techniques, live imaging of bird embryos, image analysis, and mathematical modeling, we are currently exploring different aspects of axis elongation at the multi-tissue scale. To better understand the coordination between cell specification and morphogenesis we are also studying neuro-mesodermal progenitors, a stem cell population located in the embryonic tail. Our results suggest that spatial cell-to-cell heterogeneity within progenitor cells is playing a key role in the robustness of multi-tissue morphogenetic processes.
Topologically Active Polymers
Davide Michieletto (University of Edinburgh)
Polymer physics successfully describes most of the polymeric materials that we encounter everyday. In spite of this, it heavily relies on the assumption that polymers do not change topology (or architecture) in time or that, if they do alter their morphology, they do so in equilibrium. This assumption spectacularly fails for DNA in vivo, which is constantly topologically re-arranged by ATP-consuming proteins within the cell nucleus.
Inspired by this, here I will discuss some non-equilibrium complex fluids in which DNA topology is selectively altered in time by proteins. I argue that solutions of "topologically active” polymers can display unconventional viscoelastic behaviours and can be conveniently realised using solutions of DNA functionalised by certain families of vitally important proteins.
More specifically, in this talk I will present some results using both theory/simulations and experiments on the microrheology of entangled DNA undergoing digestion by restriction enzymes and ligation by ligase. I will try to convince you that that we can harness these topological processes to design complex fluids with exotic and time-varying viscoelastic behaviours.
Robustness in miRNA-based circuits
Silvia Grigolon (Laboratoire Jean Perrin, Sorbonne U.)
Phenotypic differentiation often relies on bimodal distributions of gene expression levels, which can normally be achieved by different molecular mechanisms. During the past decades microRNAs, small noncoding RNA molecules, were found to downregulate the expression of preferred mRNA targets by sequestering and successively degrading them, thus influencing the level of gene expression. In this talk, I will show how miRNAs play an important role in filtering noise in gene expression. Starting from purely intrinsic noise, I will talk about how this can be buffered by miRNAs, which yet become apparently inefficient in the presence of sources of extrinsic noise. I will then theoretically address the question on how microRNA-mediated regulation can induce the appearance of bimodal phenotypes. Our findings show that the presence of extrinsic noise favours bimodal distributions. This suggests a simple mechanism for obtaining bimodal populations where the presence of extrinsic noise relaxes the requirements on parameters fine tuning.
Fluid structure interaction of a microcapsule in flow: application to the characterization, enrichment and sorting of capsule suspensions
Anne-Virgine Salsac (Université de Technologie de Compiègne)
Encapsulation consists in enclosing an internal medium in a solid semi-permeable membrane to protect it and control the exchanges with the environment. Being at the source of innovative applications in the fields of biotechnologies, pharmacology, or food industry, capsules offer tremendous potential in the process engineering world. But scientific challenges remain to be met, such as finding the optimal compromise between payload and membrane thickness, characterizing the membrane resistance and controlling the moment of rupture.
We will explore the challenges to use deformable liquid-core capsules of micrometric size to efficiently transport active material, with a primary focus on health-related applications. Being used suspended in a carrying fluid in flow, microcapsules constitute a formidable problem of complex fluid-structure interactions. I will briefly present how the three-dimensional capsule-flow interactions may be modeled and how these sophisticated numerical models can dialogue with microfluidic experimentations to produce innovative techniques to characterize the mechanical properties of deformable capsules, sort them upon their rigidity or enrich suspensions.
Biosketch
After graduating from University of California San Diego (USA) in 2005 with a PhD in Biofluids, Anne-Virginie Salsac was recruited as Lecturer in the Department of Mechanical Engineering of University College London (UK). In 2007, she became Researcher at the CNRS (Centre National de Recherche Scientifique). She joined the Biomechanics and Bioengineering Laboratory in Compiègne (France), where she conducts original experimental and numerical modeling of blood flows, from the microcirculation to the hemodynamics in large vessels. In particular, she focuses on the behavior of bioartificial or natural capsules in micro-channels and on the influence of innovative vascular therapeutic techniques on blood flow.
She is at the head of the Biological Fluid Structure Interaction research group since 2011, and is Visiting Professor at the Institute of Bioengineering, Queen Mary University of London (England) since 2014. She has been awarded various prices, including the CNRS bronze medal, Trophées des Femmes en Or in 2015 and Medal of the National Order of Merit in 2016, in recognition for her pluridisciplinary work.
Active-gel Theory for Multicellular Migration in the Extra-cellular Matrix
Ram Adar (Collège de France)
We formulate an active-gel theory for multicellular migration in the extracellular matrix (ECM). The cells are modeled as an active, polar solvent, and the ECM as a viscoelastic solid. Our theory enables to analyze the dynamic reciprocity between the migrating cells and their environment in terms of distinct relative forces and alignment mechanisms. We analyze the linear stability of polar cells migrating homogeneously in the ECM. Our theory predicts that, as a consequence of cell-matrix alignment, contractile cells migrate homogeneously for small wave vectors, while sufficiently extensile cells migrate in domains. Homogeneous cell migration of both extensile and contractile cells may be unstable for larger wave vectors, due to active forces and the alignment of cells with their concentration gradient. These mechanisms are stabilized by cellular alignment to the migration flow and matrix stiffness. They are expected to be suppressed entirely for rigid matrices with elastic moduli of order 10 kPa. Our theory should be useful in analyzing multicellular migration and ECM patterning at the mesoscopic scale.
Understanding protein phase transitions for globular, membrane and de novo proteins
Jennifer McManus (University of Bristol)
Protein phase transitions in biology are associated with disease pathogenesis, for example in cataract disease or sickle cell anaemia, but they also occur as part of normal biological processes, such as liquid-liquid phase separation in cells. While protein phase behaviour has been understood from a physics perspective for globular proteins for some time, how we can apply what we already know to biomolecular condensation, or the phase transitions of other protein types is less clear. In my talk, I’ll briefly review what we understand about the phase transitions of globular proteins and then present recent results on the phase behaviour of membrane proteins and the rational design of de novo proteins to undergo liquid-liquid phase separation in cells. I’ll highlight what common features exist between these different protein types and how this might be important for biomolecular condensation in cells.
Bio
Jennifer McManus completed her BSc and PhD degrees in Chemistry at University College Dublin. Following postdoctoral appointments at the University of Fribourg in Switzerland and the Massachusetts Institute of Technology, she established the Soft Matter and Biophysical Chemistry research group at Maynooth University in Ireland as a Science Foundation Ireland Stokes Lecturer. She was promoted to Associate Professor in 2016 and served as Head of Department between 2017 and 2020. She moved to the School of Physics at the University of Bristol in February 2020.Time persistence as a source of large-scale fluctuations in growing tissues
Antoine Fruleux (LPTMS)
Due to the current health situation, this week's seminar will be held online only:
https://cnrs.zoom.us/j/96254507481?pwd=VHd4SzFSUzhXZ2hJOWJ4ajhza2dmdz09
Meeting ID: 962 5450 7481
Passcode: physbi0
In biological tissues, the space and time variations of certain properties appear greatly random. Many recent studies investigate the impact of this randomness on the development of tissues. Such studies aim, for example, at understanding its role in the self-organization of tissues or at describing how the cellular responses it induces impact the developments of organs. To better understand the role played by random fluctuations during morphogenesis, a first step is to have a better characterization of their properties. Using theoretical modeling, we predicted that the spatial extend of fluctuations is large and results from the combined effects of growth and the fluctuation's time persistence. This prediction, which generally applies to any expending media, has strong consequences when considering biological tissues. It notably implies long ranged space correlations, which tend to give rise to random organ shapes, in contradiction with the observed robustness of many organ shapes. An explanation for this apparent paradox may be found in the fine tuning of some of the tissue properties to minimize the heterogeneity of organs, and we investigated the influence of the tissue's response to the mechanical stress on the organ shape. Our model’s predictions prompted us to analyze the spectra of fluctuations in growing tissues, developing a novel approach that is applicable to geometrically disordered materials. After introducing the main predictions of the model, I will present the experimental evidences supporting them in plant tissues. Finally, I will express the questions raised by our findings.
Quantifying memory effects in random search processes
Raphaël Voituriez (Laboratoire Jean Perrin)
A general question that arises in random walk theory is the quantification of space exploration by a random walker. A key observable is provided by the first-passage time, which quantifies the kinetics of general target search problems, and as such has a broad range of applications from diffusion limited reactions at the molecular scale, to immune cells patrolling tissues to find antigens, or larger scale organisms looking for ressources.
I will present asymptotic results which enable the determination of the first-passage time statistics to a target site for a wide range of random processes, and show how these results generalize to non Markovian processes, which are needed to model non Brownian, complex searchers with memory skills. I will discuss how these results can be used to assess the optimality of general random search processes. An explicit example of cellular system where long range memory effects emerge will be given.
Collective amoeboid migration of cancer cell clusters by polarised jiggling
Diane-Laure Pagès (Institut Gustave Roussy)
Winner of the PhysBio2021 best short talk award
Migration is a key step in many biological processes, including the metastatic progression of cancers which accounts for most patient’s deaths. As far as we know, cell locomotion occurs through three distinct mechanisms. In a few words, single cells can migrate via two modes, mesenchymal (adhesive, traction-based) or amoeboid (non-adhesive, propulsion-based). Cell cohorts are generally led by protrusive leaders, towing the collective through adhesion to the substrate.
We have been able to demonstrate the existence of an undescribed mode of collective migration. We study tumour cell clusters’ migration, transformed and non-transformed, in non-adherent microfabricated channels. This collective migration is independent of focal-adhesions and traction but is dependent on integrin-mediated friction to the substrate. Moreover, cell clusters display an actomyosin cortex that is polarised to the rear of clusters, proportionally to migration speed. Inhibiting ROCK and myosin activity decreases migration, while optogenetic activation of RhoA dictates directionality, demonstrating that this migration relies on actomyosin contractility [2]. However, such migration is not driven by a sustained cell or myosin flow. Instead, we observed fluctuating cell and myosin displacements that are correlated with clusters’ speed. We then demonstrate analytically that, together with friction with the substrate and myosin polarisation, this behaviour leads to migration. Our results suggest that cell clusters can use a unique mode of collective migration, based on “polarised jiggling”, that may explain the metastatic potential of these tumour intermediates. We call this new mode of migration “collective amoeboid migration”, by analogy with single cell amoeboid migration.
1. Zajac, O. et al. Tumour spheres with inverted polarity drive the formation of peritoneal metastases in patients with hypermethylated colorectal carcinomas. Nat. Cell Biol. 1 (2018). doi:10.1038/s41556-017-0027-6
2. Pagès, D.-L. et al. Cell clusters adopt a collective amoeboid mode of migration in confined non-adhesive environments. bioRxiv 2020.05.28.106203 (2020) doi:10.1101/2020.05.28.106203.
Interactions between viral factories and cellular innate immunity, a story of liquid biocondensates
Yves Gaudin (I2BC)
Replication of Mononegavirales (MNV) occurs in viral factories which form inclusions in the host-cell cytoplasm. For rabies virus (RABV), those inclusions are called Negri Bodies (NBs). NBs have characteristics similar to those of liquid organelles: they are spherical, they fuse to form larger structures, and they disappear upon hypotonic shock. Their liquid phase was confirmed by FRAP experiments. Live-cell imaging indicates that viral nucleocapsids are ejected from NBs and transported along microtubules to form either new virions or secondary viral factories.
We developed several minimal systems (both cellular and acellular) allowing the formation of biomolecular condensates recapitulating NBs properties. Those minimal systems established RABV phosphoprotein (P) as the main regulator of the liquid liquid phase separation (LLPS) and identified structural elements of RABV nucleoprotein and P that are key in this process. Formation of liquid viral factories by LLPS has been extended to other MNV. This is a paradigm change in the field of MNV replication that invites us to revisit the interplay between viral factories and innate cellular immunity.
As an example, we previously demonstrated that stress granules, which are also liquid biomolecular condensates containing microbes-associated-molecular-patterns recognition receptors acting as sensors of RNA virus replication, come into close contact with NBs, exchange material with them, but do not fully mix their content. We have now extended those observations to other components of the cellular pathway leading to interferon production demonstrating a key role of viral and cellular biomolecular condensates in innate immunity.
Elasticity of dense actin networks produces nanonewton protrusive forces at macrophage podosomes
Renaud Poincloux (IPBS, Toulouse)
Actin filaments assemble into force-generating systems that play pivotal roles in diverse cellular functions, including cell motility, adhesion, contractility and division. Thermodynamics and in vitro experiments showed that the polymerization of single actin filaments generates forces in the 1-10 pN range. How networks of crosslinked actin filaments, individually generating piconewton forces, are able to produce forces reaching tens of nanonewtons remains unclear. We used in situ cryo-electron tomography to unveil how the nanoscale architecture of macrophage podosomes enables basal membrane protrusion. We show that the sum of the actin polymerization forces at the membrane is not sufficient to explain the protrusive forces generated by podosomes. Quantitative analysis of podosome organization demonstrates that the core is composed of a dense network of bent actin filaments storing elastic energy. Theoretical modelling of the network as a spring-loaded elastic material reveals that it exerts forces of up to tens of nanonewtons, similar to those evaluated experimentally. Thus, taking into account not only the interface with the membrane but also the bulk of the network is crucial to understand force generation by actin machineries. Our integrative approach sheds light on the elastic behavior of dense actin networks and opens new avenues to understand force production inside cells.
Structure and assembly of single intermediate filaments
Cécile Leduc (Institut Jacques Monod)
Intermediate filaments (IF) are involved in key cellular functions including polarization, migration, and protection against large deformations. These functions are related to their remarkable ability to extend without breaking, a capacity that should be determined by the molecular organization of subunits within filaments. However, this structure-mechanics relationship remains poorly understood at the molecular level. Here, using super-resolution microscopy (SRM), we show that vimentin filaments exhibit a ~49 nm axial repeat both in cells and in vitro. As unit-length-filaments (ULFs) precursors were measured at ~59 nm, this demonstrates a partial overlap of ULFs during filament assembly. Using an SRM-compatible stretching device, we also provide evidence that the extensibility of vimentin is due to the unfolding of its subunits and not to their sliding, thus establishing a direct link between the structural organization and its mechanical properties. Overall, our results pave the way for future studies of IF assembly, mechanical and structural properties in cells.
SEMINAR CANCELLED
Marion Jasnin (MPI for Biochemistry, Martinsried, Germany)
Prion replication and structural diversification mechanisms
Davy Martin (INRA Jouy-en-Josas)
Prions are proteinaceous infectious agents responsible for a range of neurodegenerative diseases in animals and humans. Prion particles are assemblies formed from a misfolded, β-sheet rich, aggregation-prone isoform (PrPSc) of the host-encoded cellular prion protein (PrPC). Prions replicate by recruiting and converting PrPC into PrPSc. PrPSc is a pleiomorphic protein as different conformations can dictate different disease phenotypes in the same host species. This is the basis of the strain phenomenon in prion diseases. Moreover, recent experimental evidence suggests further structural heterogeneity in PrPSc assemblies within specific prion populations and strains. Still, this diversity is rather seen as a size continuum of assemblies with the same core structure, while analysis of the available experimental data points to the existence of structurally distinct arrangements. The atomic structure of PrPSc has not been elucidated so far, making the underlying mechanistical processes of emergence and coevolution of structurally distinct assemblies difficult to understood. Here, we will present our latest experimental results and replication model providing mechanistic insights into prion structural diversification, a key determinant for prion adaptation and toxicity.SEMINAR CANCELLED
Anne-Virginie Salsac (Université de Technologie de Compiègne)
Native membrane visualization by meta-replica electron microscopy
Stéphane Vassilopoulos (Sorbonne Université)
In cell biology, particular Importance is given to developing new methods of sample preparation that will achieve a more natural appearance of samples in the microscope. To accomplish this, scientists have developed the metal-replica electron microscopy (EM) technique directly on unroofed cultured cells. This particular EM technique permits visualization of the inner side of cell membranes and their components with an extremely high resolution using transmission EM. Metal-replicas have profoundly shaped our understanding of the functional morphology of the cell and used to capture several different cellular processes including remodeling of cytoskeletons, membrane trafficking, cell-to-cell contacts and formation of extracellular matrices. During this presentation, I will outline the history of this particular EM technique and describe different methodologies and key applications in cell biology with a particular emphasis on visualization of actin filament networks at the cell cortex. Selected publications: Vassilopoulos S*, Gibaud S, Jimenez A, Caillol G, Leterrier C. Ultrastructure of the axonal periodic scaffold reveals a braid-like organization of actin rings. Nat Commun. 2019 Dec 20;10(1):5803. Ferrari R*, Martin G, Tagit O, Guichard A, Cambi A, Voituriez R, Vassilopoulos S*, Chavrier P*. MT1-MMP directs force-producing proteolytic contacts that drive tumor cell invasion. Nat Commun. 2019 Oct 25;10(1):4886. Franck A, Lainé J, Moulay G, Lemerle E, Trichet M, Gentil C, Benkhelifa-Ziyyat S, Lacène E, Bui MT, Brochier G, Guicheney P, Romero N, Bitoun M, Vassilopoulos S*. Clathrin plaques and associated actin anchor intermediate filaments in skeletal muscle. Mol Biol Cell. 2019 Mar 1;30(5):579-590. Randrianarison-Huetz V, Papaefthymiou A, Herledan G, Noviello C, Faradova U, Collard L, Pincini A, Schol E, Decaux JF, Maire P, Vassilopoulos S, Sotiropoulos A. Srf controls satellite cell fusion through the maintenance of actin architecture. J Cell Biol. 2018 Feb 5;217(2):685-700. Elkhatib N, Bresteau E, Baschieri F, Rioja AL, van Niel G, Vassilopoulos S, Montagnac G. Tubular clathrin/AP-2 lattices pinch collagen fibers to support 3D cell migration. Science. 2017 Jun 16;356(6343). Vassilopoulos S*, Gentil C, Lainé J, Buclez PO, Franck A, Ferry A, Précigout G, Roth R, Heuser JE, Brodsky FM, Garcia L, Bonne G, Voit T, Piétri-Rouxel F, Bitoun M. Actin scaffolding by clathrin heavy chain is required for skeletal muscle sarcomere organization. J Cell Biol. 2014 May 12;205(3):377-93.Prion-like propagation of alpha-synuclein assemblies in synucleinopathies, similarities with Tau in Tauopathies
Ronald Melki (Institut François Jacob, CEA & CNRS)
The aggregation of proteins within the central nervous system is deleterious and associated to neurodegenerative disorders. The aggregation of the proteins alpha-synuclein and Tau are associated to synucleinopathies, in particular Parkinson's disease, and tauopathies, among which Alzheimer’s disease, respectively. How alpha-synuclein and Tau aggregates, how those aggregates traffic between cells, amplify by recruiting endogenous monomeric alpha-synuclein or Tau and cause distinct synucleinopathies or tauopathies is unclear. I will explain the molecular events that lead to alpha-synuclein or Tau aggregation. I will show that alpha-synuclein and Tau aggregates bind to neurons cell membranes and explain the cellular consequences of binding. The similarities and differences between alpha-synuclein and Tau will be highlighted. I will explain how alpha-synuclein and Tau aggregates penetrate the cells and get transported. Finally, I will describe how the structure of the fibrillar polymorphs alpha-synuclein and Tau aggregation yield distinct diseases.References:
- Brundin P et al. (2010) Nat Rev Mol Cell Biol. 11:301-7.
- Bousset L et al. (2013) Nat Commun. 4:2575.
- Peelaerts W et al. (2015) Nature 522:340-4.
- Shrivastava AN et al. (2015) EMBO J. 34:2408-23.
- Brahic M et al. (2016) Acta Neuropathol. 131:539-48.
- Makky A et al. (2016) Sci Rep. 6:37970.
- Flavin W et al. (2017) Acta Neuropathol. 134:629-653.
- Shrivastava AN et al. (2017) Neuron 95:33-50.
- Melki R (2018) Neurobiol Dis. 109:201-208.
- Gribaudo S et al. (2019) Stem Cell Reports. 12:230-244.
- Shrivastava AN et al. (2019) EMBO J. 38. pii: e99871.
- Fenyi A et al., (2019) Neurobiol Dis 129:38-43.
A new approach unifies mechanical rigidity in cell-based tissue models and biopolymer networks
Matthias Merkel (Turing Center for Living Systems, Marseille)
Understanding how mechanical properties of biological tissues arise from collective cellular behavior is vital for understanding the mechanisms that guide embryonic development, cancer growth, and wound healing. With my group, I am studying several questions of collective effects and self-organization in biological tissue. Recently, a new type of rigidity transition was discovered in a family of cell-based models for 2D and 3D tissues. Here I discuss these transitions and show that they are an instance of a much more general class of transitions, which appear when introducing geometric incompatibility into so-called under-constrained systems. This kind of transition also provides an important limiting case to understand stiffening in fiber network models, which are used to describe biopolymer networks like collagen. We show that all of these models exhibit generic elastic behavior close to the transition, which is largely independent of the microscopic structure and the disorder in the system. We obtain analytic expressions for the relevant elastic properties and numerically verify our findings by simulations of under-constrained spring networks as well as 2D and 3D vertex models for dense biological tissues. Several of our predictions are parameter-free, and we thus expect them to be general hallmarks for geometric-incompatibility-induced stiffening in under-constrained materials. Hence, they provide quantitative experimental tests for whether stiffening in a given material is due to this effect or not. Finally, I will briefly discuss another project of current interest in my group, where we will explore conditions for robust self-organized oriented deformation of biological tissue.
Probing proteins in small volumes
Tuomas Knowles (Cambridge University, UK)
Warning: special timeThis talk outlines our efforts on exploring experimental strategies to provide a new window into protein self-assembly that are enabled by operation in small volumes. We have shown that microconfinement achieved through droplet microfluidics allows the isolation of single nucleation events in protein aggregation and thus to study a rare event as single molecule resolution. Using this strategy we have also been able to develop an understanding of how aberrant misfolded protein states are transmitted from one molecule to another through time and space. More recently we have exploited measurements of mass transport through fluid streams under laminar flow conditions to generate a platform for probing protein-protein interactions under fully native conditions.
Biophysics of Killing – Theory and Experiment
Heiko Rieger (Saarland University)
Cytotoxic T lymphocytes and natural killer cells are the main cytotoxic killer cells of the human body to eliminate pathogen-infected or tumorigenic cells. Various processes are involved in a successful killing event: activation of the killer cell, migration and search for the target, formation of a synapse and polarization upon contact with the target, transport of cytotoxic agents towards the synapse, and finally elimination of the target via necrosis or apoptosis. In this talk I will review various biophysical aspects of killing that were studied in collaboration with experimental groups from biology and medicine. Topics include the analysis of search strategies of migrating killer cells; the mechanistic understanding of the molecular motor driven cytoskeleton rotation towards the synapse during polarization; the efficiency of the spatial organization of the cytoskeleton for search problems occurring in intra-cellular cargo transport; and the stochastic analysis of different killing strategies via inducing necrosis or apoptosis.
Collective motion in bacterial suspensions
Carine Douarche (Université Paris-Sud)
Warning: special seminar time.
Suspending highly motile Escherichia coli in a liquid lowers the viscosity of the solution at low shear rate. At higher cell concentrations, a regime of near zero viscosity can be reached. In this work, we investigate the system-size dependence of the rheological response of an E. coli suspension as a function of shear rate and bacteria concentration using a low-shear Couette rheometer. Additionally, we image the suspensions in a cone-plate rheo-imaging setup allowing direct visualisation of the collective organisation under shear. We find the flow becomes banded and viscosity decreases to near zero at a bacterial concentration close to where collective motion appears in absence of flow.
Indeed, dense suspensions of swimming bacteria display remarkable collective motion, i.e. local bacterial ordering associated with a characteristic correlation length, reminiscent of turbulent flow behaviour. Using video microscopy over large fields of view (up to 3 mm x 4 mm), and particle image velocimetry, we calculate the spatial correlation of the velocity vectors and extract a characteristic length scale. At sufficiently high bacterial concentrations, we find this length to be proportional to the smallest system size. However, the absence of saturation towards large system-sizes suggests there is no intrinsic length-scale in these dense populations of such ‘pusher-like’ swimmers.
Using microfluidics for quantitative studies of post-embryonic development in C. elegans
Wolfgang Keil (Institut Curie, Paris)
Special location and time; seminar hosted by Carsten Janke
The development of most metazoans can be divided in an early phase of embryogenesis and a subsequent phase of post-embryonic development. Developmental dynamics during the post-embryonic phase are generally much slower and often controlled by very different molecular mechanisms that, e.g., ensure tissue synchrony and integrate metabolic queues. However, obtaining long-term in-vivo quantitative imaging data post-embryonically with good statistical and cellular resolution has been highly challenging because animals must be allowed to grow, feed, and move in order to properly develop after embryogenesis. In this talk, I will discuss our recent progress in overcoming these challenges in the model organism C. elegans, using microfluidics technology. I will then outline two of our recent studies, in which quantitative in-vivo imaging data of post-embryonic development allows novel insights into mechanisms of cell-fate acquisition and the regulation of oscillatory gene expression in C. elegans.
Membrane reshaping induced by Curvature sensitive Septin filaments: A story of paired filaments told by a pair of Orsay alumni
Stéphanie Mangenot & Aurélie Bertin (Institut Curie, Paris)
Septins are cytoskeletal proteins that assemble into a variety of supramolecular organizations from paired filaments to bundles, ring like structures or gauzes of orthogonal filaments [1‐3]. Septins are bound to the inner plasma membrane through specific interactions with phosphoinositides [1,4]. Septins are essential for cell division, participate in the formation of diffusion barrier and might be involved in membrane deformation and rigidity. Throughout cell division, septins undertake major rearrangements. Septin filaments are first aligned toward the mother‐daughter cell axis and then rotate to be circumferential around the constriction site. We have shown that septins arrange differently on positive or negative curvatures using Scanning Electron Microscopy on micro‐patterned PDMS periodic undulated substrates. Besides, this curvature preference is closely related to the ability of septins to reshape and deform membranes. Indeed, bound to Giant unilamellar Vesicles (GUVs), septins induce striking deformations with regular spikes and hollow micrometric deformations at the surface of liposomes, as visualized by fluorescence microscopy. Smaller vesicles (LUVs of 100‐300 nm in diameter), highly positively curved, are flattened by Septin filaments into “pancake”like objects as shown in 3D by cryo‐electron tomography. With the resolution of cryo‐EM and sub‐tomogram averaging we visualize both the septin filaments and the deformed vesicles. We propose a simple model where the filamentous properties of septins control their curvature sensitivity and thus impose their orientation in situ [5].- [1] A. Bertin, et al. (2010), Phosphatydinositol 4,5 biphosphate promotes budding yeast septin filament assembly and organization, J. Mol. Biol., 404(4), 711‐31.
- [2] G. Garcia et al. (2011), The regulatory budding yeast septin Shs1 promotes ring and gauze formation in a phosphorylation dependent manner., J. Cell. Biol., 195(6), 993‐1004.
- [3] A.Bertin, et al. (2008), Saccharomyces Cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly, Proc.Natl. Acac. Sci USA, 105, 8274‐8279
- [4] Beber A et al.,Septin‐based readout of PI(4,5)P2 incorporation into membranes of giant unilamellar vesicles. (2018) Cytoskeleton. doi: 10.1002/cm.21480
- [5] A. Beber et al. (2019), Membrane reshaping by micrometric curvature sensitive septin filaments, Nat. Commun., 10, 420.
Intelligence, a recipe
Matthew Turner (Warwick University, UK)
We study information-processing (artificial), or “intelligent” (living), agents. These agents seek maximal control of their environment via future state maximisation (FSM), a principle that we argue may relate to intelligent behaviour more generally. Here we study moving, re-oreintable agents that seek to maximise their space of accessible (visual) environments, out to some time horizon. The action of each agent is (re)established by exhaustive enumeration of its future decision tree at each time step - each agent chooses the branch of its tree leading from the present to the richest future state space. Remarkably, cohesive swarm-like motion emerges that is similar to that observed in animal systems, such as bird flocks. We develop heuristics that mimic computationally intensive FSM but that could also operate in real time under animal cognition. Finally, we show that iterative application of these heuristics as the model for the behaviour of others, when determining the dynamics of self under full FSM, can lead to a form of closure for the problem. I will argue that this offers a philosophically attractive, bottom-up mechanism for the emergence of swarming.
Some aspects of immune cells mechanobiology: activation of T lymphocytes and parasite phagocytosis by macrophages
Pierre-Henri Puech (Adhesion and Inflammation Lab - Aix-Marseille Universités)
Surface proteins of cells which are implicated in cell recognition and adhesion are also subjected to forces, since the interactions with their partner is often occuring at the interface between two cells. The study of the roles of physical cues, such as forces, substrate mechanics and dynamics has lead to the emergence of mechanobiology. One idea is to dissect how these physical cues "help" the cells to read and interpret, sometimes react to, biochemical messages, a process known as mechanotransduction.
In this seminar, we will present why and how forces and mechanics are thought to be key regulators of immune cell recognition using our work using two biological models:
- the crucial step of T cell activation which leads to an efficient immune response
- the phagocytic response of macrophages to the parasite Toxoplasma Gondii, responsible of toxoplasmosis.
SEMINAR CANCELLED
Stefan Karpitschka (MPI Göttingen)
Bacterial aggregates of Neisseria meningitidis flow inside blood vessels like a honey-like viscous liquid
Daria Bonazzi (Institut Pasteur, Paris)
During human meningococcal infections, bacteria accumulate and finally fill up blood vessels, causing severe diseases such as septicaemia and meningitis. However, the mechanisms governing formation of bacterial aggregates and their impact on infection remained elusive. In this multidisciplinary work we unravel that aggregates of Neisseria meningitidis behave as a viscous liquid similar to honey. This is due to type-IV pili, long adhesive filaments which are constantly extending and retracting at the bacterial surface. By these means, bacteria can find each other and transiently come into contact. Aggregation is therefore based on an intermittent process of attraction between bacteria, and gives rise to a new type of active material. Importantly, the unique physical properties of meningococcal aggregates allow them to gradually adapt to the complex geometry of the vascular network. As a consequence, these physical properties are essential for the progression of infection.
Formation of intracellular amorphous carbonates by bacteria
Karim Benzerara (Sorbonne Universités, Paris)
Living cells can sustain out-of-equilibrium states in a given environment by consuming free energy. The formation of some intracellular mineral phases provides some examples of this. Here I will review some of the work we have performed in the last years to describe and understand how some cyanobacteria, which are abundant photosynthesizing bacteria appeared several billions years ago at the surface of the Earth, manage to form intracellular amorphous carbonates. I will show the environmental conditions under which they catalyze this process, detail the methodologies (including cryo-TEM and spectroscopies) we used to characterize these phases and finally address the involved (bio)molecular mechanisms. The question of the selective advantage(s) provided by this process, if any, will be asked. The implications for basic and applied research will also be addressed. Overall, this talk should convince you that a highly interdisciplinary work is crucially needed to globally understand this intracellular biomineralization process.
Developing and applying fast constant pH methods in biological systems: From biomaterials to virus
Fernando Luis Barroso Da Silva (University of São Paulo, Brazil)
SPECIAL TIME & DATE
pH is a key parameter for biological and technological processes. Different numerical schemes were developed during the last years for such simulations ranging from Poisson-Boltzmann approaches to explicit solvent based methods. Ideally, the proton equilibria should correctly describe the experimental system without hampering the calculation time. A fast proton titration scheme (FPTS), rooted in the Kirkwood model of impenetrable spheres, where salt is treated at the Debye-Huckel level, was specially developed for proteins and nucleic acids. This method has now been coupled with OPEP5 force field for constant pH molecular dynamics simulations. A benchmark study will be presented. Despite our approximations, both the robustness and its ability to proper describe the system physics by these numerical methods can be confirmed. FPTS was also applied to quantify protein stability and biomolecular interactions. In this talk, I will present results for some protein systems with importance in different applied fields from biomaterials to public health.
Sensing and creating mechanical forces by protein assembly
Andela Saric (University College London, UK)
Responding to mechanical cues and producing mechanical forces is central to survival and adaptation of all organisms. To do so, cells dynamically organise a large number of membrane proteins into functional nanoscale structures. Due to their multiscale nature, such processes are challenging to resolve with current experimental techniques and are too complex for detailed molecular simulations. Coarse-grained computer simulations, rooted in statistical mechanics and soft-matter physics, can be of great value in determining how functional protein nanostructures operate. Here I will discuss cooperative gating of mechanosensitive membrane channels, and the production of mechanical forces by elastic filaments adsorbed on membranes, using minimal coarse-grained models. I will provide quantitative predictions for cooperative action of bacterial mechanosensitive channels, from molecular to cellular scales. I will propose how geometrical transitions of ESCRT-III filaments can remodel and sever membranes of various shapes and topologies, creating membrane cones, tubes, buds, and dividing tubular and spherical membranes. Beyond their biological context, our findings can also guide the design of artificial structures that mechanically sculpt cells and sense mechanical forces at the nanoscale.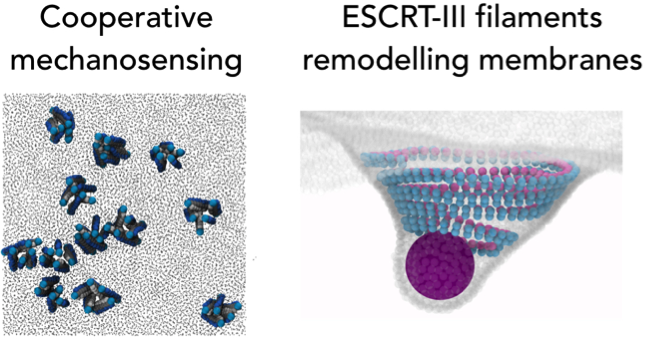
Modeling evolution of brain tumors
Mathilde Badoual (IMNC, U. Paris-Sud)
Diffuse low-grade gliomas are slowly-growing tumors. After tens of years, they transform inexorably into more aggressive forms, jeopardizing the patient’s life. Mathematical modeling could help clinicians to have a better understanding of the natural history of these tumors and their response to treatments. We present here different models of these tumors: the first one is discrete and describes the appearance of the first glioma cells and the genesis of a tumor. The second model is continuous and consists in a partial differential equation that describes the evolution of the cell density. This model can describe the natural evolution of gliomas and their response to treatments such as radiotherapy. The discrete and the continuous models are designed to be close to the biological reality. The results are quantitatively compared with either biological data or clinical data, at the cellular level (histological samples) and at the organ level (clinical imaging, such as MRI scans).
From cells to tissue: A continuum model of epithelial mechanics
Philippe Marcq (Institut Curie, Paris)
SPECIAL LOCATION
A continuum model of epithelial tissue mechanics is formulated using cellular-level mechanical ingredients and cell morphogenetic processes, including cellular shape changes and cellular rearrangements. This model incorporates stress and deformation tensors, which can be compared with experimental data. Focusing on the interplay between cell shape changes and cell rearrangements, we elucidate dynamical behavior underlying passive relaxation, active contraction-elongation, and tissue shear flow. Extensions of the model allow to incorporate additional ingredients, such as cell division, cell death, and cell motility.
Seminar cancelled: rescheduled for March 13th
Karim Benzerara (Sorbonne Universités, Paris)
Single molecule analysis of mitochondrial permeabilization in apoptosis
Ana-Jesus Garcia-Saez (MPI Tübingen, Germany)
Bax and Bak are key regulators of apoptosis and mediate the permeabilization of the outer mitocondrial membrane that leads to cytochrome and Smac release. Although it is widely accepted that Bax and Bak function and molecular mechanism largely overlap, there is limited evidence how Bak works. In previous studies, we have used single molecule microscopy to characterize the oligomerization of Bax in the membrane and its organization at the nanoscale in the mitochondria of apoptotic cells. We now extended these approaches to Bak and identified key structural differences between the two proteins that may have functional implications.
How to set the proper size and shape of metaphase spindles
Jan Brugues (MPI Dresden, Germany)
Regulation of size and growth is a fundamental problem in biology. A prominent example is the formation of the mitotic spindle, where protein concentration gradients around chromosomes are thought to regulate spindle growth by controlling microtubule nucleation. Previous evidence suggests that microtubules nucleate throughout the spindle structure. However, the mechanisms underlying microtubule nucleation and its spatial regulation are still unclear. In the first part of the talk I will present an assay based on laser ablation to directly probe microtubule nucleation events in Xenopus laevis egg extracts. Combining this method with theory and quantitative microscopy, we show that the size of a spindle is controlled by autocatalytic growth of microtubules, driven by microtubule-stimulated microtubule nucleation. The autocatalytic activity of this nucleation system is spatially regulated by the limiting amounts of active microtubule nucleators, which decrease with distance from the chromosomes. This mechanism provides an upper limit to spindle size even when resources are not limiting. Once the necessary amounts of microtubules are created, the activities of motors lead to the proper shape and architecture of spindles. In the second part of the talk I will discuss the origin of motor-mediated stress in spindles.Live imaging of motile cilia to investigate left-right symmetry breaking in zebrafish embryos
Willy Supatto (LOB, École polytechnique)
In vertebrate embryos, cilia-driven fluid flows are guiding left-right body symmetry breaking within the left-right organizer (LRO). To investigate the generation and sensing of flows, it is required to quantify cilia biophysical features in 3D and in vivo [1]. In the zebrafish embryo, the LRO is called the Kupffer’s vesicle (KV) and is a spheroid shape cavity, which is covered with motile cilia distributed at its surface and oriented in all directions of space. This transient structure varies in size and shape during development and from one embryo to the other. As a consequence, the experimental investigation of cilia properties is challenging. It requires quantifying cilia features in vivo and in 3D and combining the data from different embryos to compare one embryo to the other and perform statistical analyses.To reach this goal, we devised an experimental workflow combining live 3D imaging using multiphoton microscopy, image processing, and data registration to quantify cilia biophysical features, such as cilia density, motility, 3D orientation, or length. We integrated such experimental features obtained in vivo into a fluid dynamics model and a multiscale physical study of flow generation and detection. This strategy enabled us to demonstrate how cilia orientation pattern generates the asymmetric flow within the KV [2]. In addition, we could investigate the physical limits of flow detection to clarify which mechanisms could be reliably used for body axis symmetry breaking [2]. Finally, we discovered the distribution of cilia orientation is asymmetric within the KV [3]. Importantly, these results suggested that the asymmetric force detection could result from the cilium being sensitive to its own motion. Together, this work sheds light on the complexity of left-right symmetry breaking and chirality genesis in developing tissues.
[1] From cilia hydrodynamics to zebrafish embryonic development. Supatto & Vermot, Current Topics in Developmental Biology 2011
[2] Physical limits of flow sensing in the left-right organizer. Ferreira et al, eLife 2017
[3] Chiral cilia orientation in the left-right organizer. Ferreira et al, Cell Reports, in press
Cell contraction induces long-ranged stress stiffening in the extracellular matrix
Pierre Ronceray (Princeton University, USA)
Animal cells in tissues are supported by biopolymer matrices, which typically exhibit highly nonlinear mechanical properties. While the linear elasticity of the matrix can significantly impact cell mechanics and functionality, it remains largely unknown how cells, in turn, affect the nonlinear mechanics of their surrounding matrix. Here we show that living contractile cells are able to generate a massive stiffness gradient in three distinct 3D extracellular matrix model systems: collagen, fibrin, and Matrigel. We decipher this remarkable behavior by introducing Nonlinear Stress Inference Microscopy (NSIM), a novel technique to infer stress fields in a 3D matrix from nonlinear microrheology measurement with optical tweezers. Using NSIM and simulations, we reveal a long-ranged propagation of cell-generated stresses resulting from local filament buckling. This slow decay of stress gives rise to the large spatial extent of the observed cell-induced matrix stiffness gradient, which could form a mechanism for mechanical communication between cells.
How do cells measure their boundaries to tailor physiological responses?
Alexis Lomakin (King's College London, UK)
Much like modern day engineered devices, cells in the human body are able to make precise measurements: intestinal epithelial cells monitor local cell densities to prevent hyperplasia, neutrophils sample their microenvironment to compute the fastest migratory route toward infection sites, and epidermal stem cells use extracellular matrix occupancy to make cell fate decisions. What these examples illustrate is the sensitivity of complex cell behaviors to spatial and mechanical constraints, known in quantitative sciences as boundary conditions. Although the importance of boundary conditions in cell and tissue physiology is increasingly recognized, it remains unclear how cells sample their boundaries to tailor specific behaviors to boundary conditions. Here, using biophysical tools to manipulate cell boundaries in a highly controlled, quantitative manner, we found that cells estimate externally-imposed confinement using their largest and stiffest intracellular component, the nucleus. Cell confinement below a certain threshold deforms the nucleus and expands its envelope area. Unbuffered against area expansion due to slow turnover of constituents, the nuclear envelope becomes stretched. This in turn engages signaling via nuclear membrane stretch-sensitive proteins to the actomyosin cortex, activating contractility. The latter provides a motive force for the cell to squeeze through tight pores and constrictions in the extracellular matrix. Interestingly, no increase in cell contractility is observed when cells move through environmental confines that do not significantly deform the nucleus. Thus, the nucleus acts as an internal ruler for environmental confinement size, allowing cells to utilize energetically costly contractility on demand, only when surrounding space becomes restrictive. The advantage of the proposed mechanism is that in contrast to the plasma membrane, nuclear membranes do not participate in constitutive membrane trafficking; their surface area thus fluctuates less. This intrinsic quiescence should privilege them to function as low-noise detectors, to readily discriminate local environmental conditions from internal traffic-induced cell area/tension fluctuations.
Bacterial collective behaviours
Knut Drescher (Max Planck Institute for Terrestrial Microbiology, Marburg, Germany
In nature, bacteria often engage in a range of collective behaviors. In this presentation, I will demonstrate how two bacterial behaviors, swarming and biofilm formation, are related by physical interactions, chemical signaling, and dynamical transitions. I will show how these collective behaviors arise from cell-cell interactions, and the physiological state of individual cells. Furthermore, I will introduce new experimental methods for investigating bacterial collective behaviors.
Quantitative analysis of the somitogenetic wavefront
David Bensimon (LPS-ENS, Paris, France)
Somitogenesis is the process by which the anterio-posterior axis is segmented in all vertebrates thus defining the coordinate system that will serve for positioning of the appendices and organs. This process of segmentation is due to the interaction between a posterior moving wavefront of morphogens and a posterior located clock generating somites (segments) at regular times and places. The existence and characterization of the clock has been amply demonstrated. In this talk I will focus on the molecular network behind the wavefront. I will discuss the wavefront response to various perturbations and compare our observations with a model of this network.
Collagen and gelatin from sol to gel states for the synthesis of biomaterials
Gervaise Mosser (Laboratoire de Chimie de la Matière Condensée, Université Pierre et Marie Curie)
Collagen type I, the most abundant protein of connective tissues (bones, dermis, tendons, etc), is a macromolecular mesogen that can form lyotropic liquid-crystal phases. With this approach, our team works on elaborating several biomimetic biomaterials. However, the use of collagen can be hindered due to its price and the possibility to easily denature into gelatine and noticeably during sterilization processes. In this context, we wanted to determine whether collagen could be partially replaced by gelatine without modification of the overall hierarchical structure of the biomaterial.
Nanodiamond based quantum sensors for biological applications
Quan Li (Chinese University of Hong Kong, China)
Special location: Laboratoire Aimé Cotton, Orsay
Nanodiamond (ND) with Nitrogen-vacancy (NV) centers serves as promising bio-sensor due to its excellent bio-compatibility, high photo-stability, and the long spin coherence time at room temperature. However, the complicated biological environment, e.g. in a single cell, imposes stringent requirements on the sensor probes to be internalized. In this talk, I will discuss the requirements on nanodiamond as intra-cellular sensor, and the possible strategies that will enable various bio-sensing measurements. I will start with the understanding of nanodiamond-cell interfaces, from anchoring of ND on the plasma membrane to their internalization, and eventually to their intracellular trafficking. Other than the conventional three-dimensional trajectories tracking of the ND, it is also possible to track their orientations (rotation), providing additional information of the intracellular environment. One problem with NV based bio-sensing is that the NV center is less sensitive to certain parameters such as temperature and pressure, and not at all response to many other important biochemical parameters such as pH and non-magnetic biomolecules. I will also discuss possible schemes of constructing nanodiamond based hybrid sensors, which lead to significantly enhanced sensitivity and/or potentially enable the measurement of various biochemical parameters using NV based quantum sensing.
Plant vibration, from wind flutter to phenotyping
Emmanuel de Langre (École polytechnique, Palaiseau, France)
Plants are often very flexible objects. This results in motion under stimuli such as wind or currents, but also hosts such as insects. Motion are known to influence plant development by thigmomorphogenesis. I will review methodologies and results from the past ten years, aimed at quantifying and understanding the vibration of plants, or parts of plants, from Arabidopsis Thaliana to large trees. I will focus on experimental techniques indoor and outdoor, on simple models of motions, and the role of the plant architecture. The recent application to high throughput plant phenotyping by vibrations will also be presented.
Non-linear stimulus processing by the retina
Ulisse Ferrari (Institut de la Vision, Paris)
Understanding how sensory systems process information is an open challenge. This is mostly because these systems are non-linear, making it extremely difficult to model the relation between the stimulus and the sensory response. In this talk I will discuss two strategies to tackle this problem and apply them to the retina.
First, we use ex-vivo multi-electrode array experiments to record the retinal activity and directly model the ganglion cell response to complex stimuli, such as videos of moving objects. Here I will show that standard, nearly-linear, models are not enough and highly non-linear models are required. Then I will present the result of a closed-loop experiment where we adapted the stimulus on-line to investigate how the response changes when the visual stimulation is perturbed.
With this approach we could estimate the optimal performance of a neural decoder and show that the non-linear sensitivity of the retina is consistent with an efficient encoding of stimulus information.
References:
-) U. Ferrari, C. Gardella, T. Mora, O. Marre. eNeuro, vol. 4, 6. 2017.
-) S. Deny, U. Ferrari, P. Yger, R. Caplette, S. Picaud, G. Tkacik, O. Marre, Nature Commun. 8 (1) 2017
Mechano-sensitive adhesion in cell spreading and crawling
Pierre Sens (Institut Curie, Paris)
Crawling cell motility is powered by actin polymerization and acto-myosin contraction. When moving over a flat and rigid substrate, cells usually develop thin and broad protrusions at their front, called lamellipodia, where actin polymerisation generates a protrusive force pushing the front edge of the cell forward. The lamellipodium displays interesting dynamics, including normal and lateral waves, possibly relevant to cell polarisation and the initiation of motion. I will discuss a stochastic model of mechano-sensitive cell adhesion, and discuss its relevance for symmetry breaking, cell polarisation, and motility. I will then discuss a generic model of micro-crawlers, built as an extension of low Reynolds number micro-swimmers, that highlights the crucial role of mechano-sensitive adhesion for the active crawling of cells and biomimetic objects.
Sensing the matrix: transducing mechanical signals from integrins to the nucleus.
Pere Roca-Cusachs (Institute for Bioengineering of Catalonia, Universitat de Barcelona, Spain)
Cell proliferation and differentiation, as well as key processes in development, tumorigenesis, and wound healing, are strongly determined by the properties of the extracellular matrix (ECM), including its mechanical rigidity and the density and distribution of its ligands. In this talk, I will explain how we combine molecular biology, biophysical measurements, and theoretical modelling to understand the mechanisms by which cells sense and respond to matrix properties. I will discuss how the properties under force of integrin-ECM bonds, and of the adaptor protein talin, drive and regulate matrix sensing. I will further discuss how this sensing can be understood through a computational molecular clutch model, which can quantitatively predict the role of integrins, talin, myosin, and ECM receptors, and their effect on cell response. Finally, I will analyze how signals triggered by rigidity at cell-ECM adhesions are transmitted to the nucleus, leading to the activation of the transcriptional regulator YAP.
Adaptive division control in stressed bacterial cells
Shiladitya Banerjee (UCL, UK)
Control of cell size is a fundamental adaptive trait that underlies the coupling between cell growth and division. Cells possess the unique ability to adapt their size and shapes in response to environmental cues, thereby translating extracellular information into decisions to grow or divide. However, the physical mechanisms mediating the regulation of cell size and division timing remain poorly understood. In this talk, I will discuss our recent discovery of an adaptive model of cell size control in bacteria, where the decision to divide is tightly regulated by the spatial patterning of cell wall growth modes. Using a combination of stochastic mechanical modelling and single-cell experiments, I will elucidate the implications of the size control model for cellular fitness adaptation under stress. In particular, our results show that morphological transformations provide fitness and survival advantages to bacteria under sustained antibiotic treatment.
Controled oxidation in living systems
Stéphanie Bonneau (Laboratoire Jean Perrin, Paris)
Living systems produce energy by oxidizing carbon : in aerobic organisms, a major step of this oxidation is processed by the respiratory chain in mitochondria. Energy production involves oxidation and subsequent ageing of the cellular materials. The control of their oxidative activity allows cells to remain far enough to the thermodynamic equilibrium and consequently the balance between respiration and ageing is a major regulation parameter of cell's fate. The key role of mitochondria in this phenomenon will be discussed.
Experimentally, the control of the cellular oxidation is performed by using chosen photosensitizers. Due to their macrocycle, such molecules present very special photo-physical properties. Their light irradiation generates, through their triplet state, reactive oxygen species. The lifetime of these molecular species is very short and their action is very localized. To specifically target photosensitizers to one or the other cell compartments is thus the basis of their potential to modify and control the physiology of the cells. For example, the photo-chemical internalization (PCI) of macromolecules into cells is based on the photo-induced alteration of endosomal membranes - before their maturation in lysosomes - allowing the escape of the macromolecules, free to reach its targets within cell. More extensive photo-induced changes, in particular to the mitochondria, lead to cell death by necrosis or apoptosis. This photo-induced cell death is basis of an anticancer therapy so-called PDT.
First, we focussed on the photo-induced modifications of the cellular trafficking. By combining measurements of local cytoplasmic viscosity and active trafficking, we found that photodynamic effect induced a only slight increase in viscosity but a massive decrease in diffusion. These effects are the signature of a return to thermodynamic equilibrium of the system after photo-activation. Secondly, to better apprehend such complex effects, we turned to model systems. In particular, we focused on photo-oxidation of membranes lipids, that are important oxidative targets. We extensively studied their modifications under photo-oxidation. Our purpose is to demonstrate that the photo-induced permeabilization of the membranes is correlated with a deep physical stress, which can be relaxed by various pathways, depending on its lipids composition, which is characteristic of the targeted cellular compartment.
SEMINAR CANCELLED.
Jan Brugués (MPI-CBG Dresden)
Confining and releasing cell monolayers
Pascal Silberzan (Institut Curie, Paris)
Cell monolayers routinely exhibit collective behaviors largely controlled by cell-cell interactions. In this context, confinement and boundary conditions play an important role in the organization and dynamics of these cell assemblies. Interestingly, many in vivo processes, including morphogenesis or tumor maturation, involve small populations of cells within a spatially restricted region.
We report experiments in which epithelial monolayers confined in circular disks exhibit low-frequency periodic radial displacement modes. When the boundary is removed, cells collectively migrate on the free surface. The essential characteristics of the collective dynamics in these two situations are well-accounted for by the same theoretical model in which cells are described as persistent random walkers which adapt their motion to that of their neighbors.
In contrast, elongated fibroblasts that do not develop significant cell-cell adhesions self-organize until reaching a perfect nematic order upon confinement in linear stripes. When the cells are confined within a disk, the number and charge of the topological defects characteristic of nematics can be controlled, emphasizing the role of friction in this active nematic system.
After days in culture, the confined epithelia develop a tridimensional structure in the form of a peripheral cell cord at the domain edge. Confinement by itself is therefore sufficient to induce morphogenetic-like processes including spontaneous collective pulsations, global orientation and transition from 2D to 3D.
Molecular to cellular mechanics probed by high-speed atomic force microscopy
Felix Rico (Aix-Marseille Université)
The mechanical properties of individual proteins, filaments, and supramolecular assemblies provide structural stability and mechanical flexibility to the living cell. Thus, molecular understanding of the mechanics from the single molecule to the whole cell is relevant to understand biological function. High-speed atomic force microscopy (HS-AFM) is a unique technology that combines nanometric-imaging capabilities at video rate. In this talk, I will present our recent applications of HS-AFM to probe protein and cellular mechanics. In the first part, I will introduce the development of high-speed force spectroscopy (HS-FS) to probe protein unfolding at the timescales of molecular dynamics simulations (1). This provides a unique approach to acquire atomistic understanding of biomolecular processes based on experimental results. In the second part, I will present our recent work on the adaptation of HS-AFM to probe the microrheology of living cells at high frequencies (up to 100 kHz), revealing cytoskeletal dynamics (2). We show that the mechanical response at high frequencies depends on the actin filament tension and pathological state of the cell. Microrheology over a wide dynamic range—up to the frequency characterizing the molecular components—provides a mechanistic understanding of cell mechanics.
1. F. Rico, L. Gonzalez, I. Casuso, M. Puig-Vidal, S. Scheuring, High-Speed Force Spectroscopy Unfolds Titin at the Velocity of Molecular Dynamics Simulations. Science 342, 741 (2013).
2. A. Rigato, A. Miyagi, S. Scheuring, F. Rico, High-frequency microrheology reveals cytoskeleton dynamics in living cells. Nat Phys 13, 771 (2017).
Untangling the biological hairball of immune recognition networks
Paul François (McGill University, Canada)
Complex mathematical models of interaction networks are routinely used for prediction in systems biology. However, it is difficult to reconcile network complexities with a formal understanding of their behavior. I will introduce several models of immune recognition by T cells and will show how a simple procedure can be used to reduce them to functional submodules, using statistical mechanics of complex systems combined with a fitness-based approach inspired by in silico evolution. Our procedure works by putting parameters or combination of parameters to some asymptotic limit, while keeping (or slightly improving) the model performance, and requires parameter symmetry breaking for more complex models. An intractable model of immune recognition with close to a hundred individual transition rates is reduced to a simple two-parameter model, and connected to the ``adaptive sorting" principle that we previously identified and experimentally validated. Our procedure extracts three different mechanisms for early immune recognition, and automatically discovers similar functional modules in different models of the same process allowing for model classification and comparison.
Investigating embryogenesis using numerical simulations of biophysics
Ivo Sbalzarini (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany)
Abstract:
Development and morphogenesis of tissues, organs, and embryos emerges from the collective self-organization of cells that communicate though chemical and mechanical signals. Decisions about growth, division, and migration are taken locally by each cell based on the collective information. In this sense, a developing tissue is akin to a massively parallel computer system, where each cell or processor computes robust local decisions, integrating communication with other cells/processors. Mechanistically understanding and reprogramming this system is a grand challenge. Our vision is to develop a virtual computer model of a developing embryo, incorporating the known biochemistry and biophysics into a computational model in 3D-space and time, in order to understand the information-processing aspects of development on an algorithmic basis. While the “hardware” (proteins, lipids, etc.) and the “source code” (genome) are increasingly known, we known virtually nothing about the algorithms that this code implements on this hardware. Using examples from our work, I outline our roadmap toward a virtual embryo, and highlight challenges along the way. These range from globally optimal approaches to image analysis, to novel languages for parallel high-performance computing, to virtual reality and real-time graphics for 3D microscopy and numerical simulations of biochemical and biomechanical models. This cooperative interdisciplinary effort contributes to all involved disciplines.
Bio:
Ivo Sbalzarini is the Chair of Scientific Computing for Systems Biology on the faculty of computer science of TU Dresden, and director of the TUD-Department in the Center for Systems Biology Dresden. He also is a permanent Senior Research Group Leader with the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden. He graduated in Mechanical Engineering from ETH Zurich in 2002 (Willi Studer Award). He completed his doctorate in computer science in 2006 at ETH Zurich (Chorafas Award, Weizmann Institute of Science), where he formed a close collaboration between biology and computer science. In 2006, he was named Assistant Professor for Computational Science in the Department of Computer Science of ETH Zurich. In 2012, Ivo and his group moved to Dresden, where he became one of the founding members of the new Max Planck Center for Systems Biology and the TU-Dresden Chair of Scientific Computing for Systems Biology. He also serves as a co-leader of the biological systems path of the Center for Advancing Electronics Dresden, Dean of the International Max Planck Research School in Cell, Developmental, and Systems Biology, and Vice-Dean of the Faculty of Computer Science.
Ribosome assembly studied by single-molecule force measurements
Thierry Bizebard (IBPC, Paris)
Ribosomes belong to the most complicated structures in biology. Their assembly is a question of fundamental interest, but is still poorly understood. In vitro reconstitution studies have shown that the ribosome assembly process is highly cooperative and starts with the binding of a few ribosomal (r-) proteins to rRNA, but how these early binders act is unknown. Our work focuses on the initial phase of the assembly of the large subunit (50S) of the E. coli ribosome, which involves 23S rRNA, five r-proteins and a selection of assembly “helper” proteins. Our force measurements on single RNA molecules have allowed us to pinpoint several important properties of the early-binding r-proteins we have studied:
- These proteins bind with high cooperativity to the rRNA (as would be expected to obtain a high yield of fully assembled and active ribosomes).- The r-proteins act as molecular clamps, stabilising the RNA 3D structure.
- As such, they afford a strong mechanical and energetical stabilisation of the ribonucleoprotein structure (which is also probably necessary for optimum activity).
In the near future, we intend to further improve the potential of our single-molecule measurements by implementing combined force/fluorescence manipulations, and apply this methodology to our study of the early phase of E. coli large ribosomal subunit assembly.
Single-cell leukocyte mechanics: force generation, viscoelasticity, and rupture mechanics
Julien Husson (LadHyX, École polytechnique, France)
Leukocytes are very soft cells that perform many diverse functions: they adhere, crawl, transmigrate, kill, phagocytose or interact with other cells. During their activation, leukocytes both generate mechanical forces and change their viscoelastic properties (i.e. they stiffen/soften, get more or less viscous). We have developed micropipette-based setups to quantify single-leukocyte mechanical properties and monitor them over time while a leukocytes gets activated by a relevant stimulus. We further quantify rupture properties of cell membrane, as these help us to better understand cell structure and dynamics. We use this approach in diverse contexts involving leukocytes: activation of T lymphocytes, phagocytosis of a target by a neutrophil, or transmigration of a lymphoblast across an endothelial monolayer. We perform microrheology experiments with a profile microindentation setup [1,2], measure forces generated by T lymphocytes [3,4], characterize cell-substrate adhesion [5] or establish a rupture criteria for membrane rupture [2,6] (Figure 1). These mechanical measurements shed a new light on how cell mechanical properties evolve over a short period of time (seconds), how they adapt to the stiffness of their environment, and how intracellular signaling is involved. T-lymphocytes in the human body routinely undergo large deformations, both passively when going through narrow capillaries and actively when transmigrating across endothelial cells or squeezing through tissue. In this artistic rendering, a T-lymphocyte is aspirated in a micropipette to mimic passive deformations that occur when squeezing through narrow capillaries. The fluorescent signal is due to the entry of propidium iodide into the cell and indicates membrane rupture (Image: Julien Husson, LadHyX, Ecole polytechnique, cellmechanics.jimdo.com/galleries)
T-lymphocytes in the human body routinely undergo large deformations, both passively when going through narrow capillaries and actively when transmigrating across endothelial cells or squeezing through tissue. In this artistic rendering, a T-lymphocyte is aspirated in a micropipette to mimic passive deformations that occur when squeezing through narrow capillaries. The fluorescent signal is due to the entry of propidium iodide into the cell and indicates membrane rupture (Image: Julien Husson, LadHyX, Ecole polytechnique, cellmechanics.jimdo.com/galleries)
Whole-brain imaging during vestibular stimulation in zebrafish with a novel rotatable light-sheet microscope
Volker Bormuth (Laboratoire Jean Perrin, Université Pierre et Marie Curie)
Light-sheet microscopy allows cell resolved whole-brain calcium imaging at several brain scans per second in zebrafish larvae. Currently this technique is not compatible with dynamic stimulation of the vestibular system. We developed an ultra-stable miniaturized light-sheet microscope that can be rotated while performing whole-brain recordings. Rotating the microscope rotates the fish and stimulates the vestibular system while imaging always the same plane in the brain. We demonstrate volumetric whole-brain neuronal activity recordings during vestibular stimulation. We mapped the brain activity with cellular resolution of the vestibule-ocular reflex (VOR) which drives compensatory eyes movements to maintain clear vision during body rotation. Our long-term goal is study with this system multisensory signal processing by the vertebrate brain by combining visual with vestibular stimuli.
Molecular chaperones as cellular non-equilibrium machines.
Alessandro Barducci (Centre de Biochimie Structurale-INSERM, Montpellier)
Molecular chaperones are a vast class of proteins that maintain protein homeostasis in the cell and are thus essential for cell viability. In order to assist protein folding and prevent misfolding, most chaperones proceed through conformational cycles that are regulated by complex interaction networks and fueled by ATP-hydrolysis. A remarkable example are the 70-kDalton heat shock proteins (Hsp70s), which are essential in prokaryotes and eukaryotes and are involved in co-translational folding, refolding of misfolded and aggregated proteins, protein translocation, and protein degradation. While the investigation of Hsp70 cycle has attracted great attention in the last decades, the actual role of ATP-hydrolysis and, thus of energy consumption, in the chaperone function has been long unaddressed. Here we will prove how biochemical data, recent single-molecule fluorescence experiments and molecular simulations can be combined into an appropriate theoretical framework to show that: i) ATP hydrolysis allows Hsp70 chaperones to increase their affinity for the client proteins beyond the bounds imposed by equilibrium thermodynamics ii) This ultra-affinity can be exploited to perform mechanical work on client proteins thus avoiding the formation of misfolded and potentially cytotoxic species.
Block Copolymer Assemblies Beneath the Surface: Modeling Intra-Domain Textures and Chirality Transfer to Mesodomains
Greg Grason (U. of Massachussets, Amherst)
This seminar replaces that of Thierry Bizebard, was rescheduled to May 31st. Please note the more soft matter focus. Self-assembled block copolymer (BCP) melts are a chemically-versatile platform for generating a rich spectrum of periodically-ordered nanostructures of various morphologies, from arrays of layers and columns to cubic arrays of spheres and bicontinuous networks. They are also a model system for understanding processes and properties of self-assemblies, more broadly. Decades of study of BCP assembly have uncovered the principles that connect molecular BCP structure to the translational order of the (scalar) composition profiles in the ordered states. In this talk, I will describe recent efforts to understand the generic, yet poorly known, patterns of orientational ordering of constituent chain segments that underlie the otherwise well-known “standard” BCP morphologies1. From generic properties of the random-walk statistics in BCPs, we show that the direction and degree of alignment of segments varies non-trivially from place-to-place in self-organized domains, and from one morphology to another, leading to new opportunities to manipulate and harness the physics sub-domain textures. Specifically, I will discuss how our efforts to model chiral BCPs2 which have been observed to transfer handedness of chain chemistry to the chiral symmetry of mesodomain shapes that are not formed in achiral BCPs. Our generalized orientational self-consistent field (oSCF) theory framework3 shows that the thermodynamic drive for twisted, or cholesteric, packings of segments of chiral blocks stabilizes observed helical cylinder morphologies, and suggests new mechanisms for driving formation as of yet, unobserved mesochiral domain symmetries4. References- I. Prasad, Y. Seo, L. Hall and G. M. Grason arxiv.org/1612.07994 (2016)
- G. M. Grason ACS MacroLetters 4, 526 (2015). Front Cover Story
- W. Zhao, T. Russell and G. M. Grason, J. Chem. Phys. 137, 104911 (2012).
- W. Zhao, T. Russell and G. M. Grason, Phys. Rev. Lett. 110, 058301 (2013).
Physical biology of chromatin: understanding the functional role of 3D chromosome folding using polymer physics
Daniel Jost (Université Grenoble Alpes)
Cellular differentiation occurs during the development of multicellular organisms and leads to the formation of many different tissues where gene expression is modulated without modification of the genetic information. These modulations are in part encoded by chromatin-associated proteins or biochemical tags that are set down at the chromatin level directly on DNA or on histone tails. These markers are directly or indirectly involved in the local organization and structure of the chromatin fiber, and therefore may modulate the accessibility of DNA to transcription factors or enzymatic complexes, playing a fundamental role in the transcriptional regulation of gene expression. Propagation, maintenance and inheritance of these epigenetic marks are crucial mechanisms in development, phenotype stabilization and disease. Experimental evidence have shown that the pattern of chromatin markers along chromosomes is strongly correlated with the 3D chromatin organization inside the nucleus. This suggests a coupling between epigenomic information and large-scale chromatin structure. Here, I will discuss our recent works using polymer physics and numerical simulations trying to understand the basic principles behind such coupling and and to propose possible functional roles for the 3D organization of chromosomes.
Enzyme clustering can induce metabolic channeling
Michele Castellana (Institut Curie, Paris)
Direct channeling of intermediates via a physical tunnel between enzyme active sites is an established mechanism to improve the efficiency of metabolic pathways. In this seminar, I will present a theoretical model which demonstrates that coclustering multiple enzymes into proximity can yield the full efficiency benefits of direct channeling. The model predicts the separation and size of coclusters that maximize metabolic efficiency, and this prediction is in agreement with the inter-cluster spacing in yeast and mammalian cells. In addition, the model predicts that enzyme agglomerates can regulate steady-state flux division at metabolic branch points: we experimentally test this prediction for a fundamental branch point in Escherichia coli bacterium, and the results confirm that enzyme colocalization within an agglomerate can accelerate the processing of a shared intermediate by one branch. Our studies establish a quantitative framework to understand coclustering-mediated metabolic channeling, as well as its application to efficiency improvement and metabolic regulation.
Building and disassembling actin filaments with proteins and forces
Antoine Jégou (Institut Jacques Monod, France)
The actin cytoskeleton comprises several networks essential for the cell to perform many key functions (motility, cell division, tissue cohesion, …). Their assembly and disassembly is tightly regulated, in space and time, by a myriad of actin binding proteins but also by the mechanical stress applied to those networks. We take advantage of a simple setup based on microfluidics and fluorescence microscopy, to manipulate actin filaments in vitro and assay the regulation of actin assembly. Focusing first on the assembly of filaments by formins, which are able to track filament barbed ends and accelerate their elongation from profilin-actin, we will show how tracking and rapid elongation are modulated by filament tension and regulatory proteins. We will then focus on ADF/cofilin isoforms, which play a central in filament disassembly. We will show how ADF/cofilin fragments and depolymerizes filaments through different mechanisms, targeting both the side and the ends of the filaments.
Deciphering the family of immune cells at the single cell level
Leïla Perié (Institut Curie, Paris)
How heterogeneous systems of cells constituting multicellular organisms establish, organize and achieve coordination persists as a central question in natural sciences. Whereas stochastic gene or protein expressions have clearly demonstrated their role in cellular heterogeneity and are widely studied (Wang and Bodovitz, 2010), the role of cell heterogeneity in the organization of multicellular organisms has been less interrogated. Addressing this question requires adequate tools that quantitatively study ensembles of cells individually rather than group of cells.
My research aims at addressing cell heterogeneity in dynamical and complex systems of cells using the hematopoietic system as a model of study. Strikingly hematopoietic cells (immune cells, platelets and red blood cells) compose over 90% of total human cells and correspond to approximately ten trillions of cells (Sender R, 2016). More importantly they all originate from the same cells, the hematopoietic stem cells (HSC), through a process called hematopoiesis. In addition, as immune and blood cells have a short life span (from hours to months) and can response to perturbations like infections, this process is highly dynamical. It is therefore an interesting and challenging model to study differentiation in a complex system at the single cell level.
To achieve this, Leïla Perié’s lab combines different experimental and mathematical/computational approaches of single cell tracing to study hematopoiesis in vivo. For example cellular barcoding is one of the lineage tracing approaches used by the Perié’s lab. It simultaneously traces the in vivo differentiation of individual cells, allowing to reconstitute the relationship between cell lineages with single cell resolution. In this seminar, we will discuss some of our recent results using cellular barcoding in hematopoiesis.
Mechanosensing: insights from experimental physics at the single-cell scale
Atef Asnacios (MSC, Université Paris-Diderot, France)
NOTE THE NEW LOCATION (due to renovation work at LPS)
As part of their physiological functions, most cells need to adapt to their mechanical environment. In particular, the rigidity of the extracellular matrix was shown to control cell traction forces, shape, and ultimately cell differentiation. In this context, most studies focused on the role of biochemical regulation in rigidity sensing.
In contrast to this biochemical signaling-centered approach, our aim is to reveal the physical/mechanical phenomena involved in mechanosensing. To this end, we have developed original single-cell techniques combining mechanical measurements (traction force, mechanical power…) with monitoring of cell structures (evanescent wave microscopy). In particular, we have designed a unique protocol allowing us to change the effective stiffness felt by a single cell in real time (~0.1 second), thus allowing us to show that an early purely mechanical response of single cell to stiffness indeed does exist.
We will present the results of experiments combining single-cell traction force measurements, dynamic control of stiffness, and monitoring of adhesion complexes, and will discuss how cell shape (~ contact angle) and mechanical adaptation to rigidity (~ impedance matching) could control cell fate.
High-fidelity computational modelling of biofilms and fibre networks
David Head (University of Leeds, UK)
NOTE THE NEW LOCATION (due to renovation work at LPS)
Mathematical models of complex systems can help develop fundamental insight and accelerate the development of novel materials and therapeutic treatments, but the simplifications that must necessarily be made reduce both validity and predictability, even for complex models that are solved numerically. Here I will describe recent results in two research streams aimed at developing and validating high-fidelity in silico models for biofilms and fibre networks, aiming to reduce the realism gap to experimental systems while maintaining sufficient performance to permit solution for times and system sizes of interest. The first is a bespoke model for biofilms in which mechanical relaxation between cells replaces the "pushing" rules typically used, and has been applied to the archetypal biofilm of dental plaque, revealing insight into long-term ecological dynamics not easily assayed experimentally. Secondly, I will discuss dynamic simulations of peptide gels and collagen hydrogel scaffolds, both having important applications as novel biomaterials but where network formation must also be simulated to better approximate the experiments; reaching realistic time scales is a substantial challenge.
Deficient ribosome biogenesis is an early marker of cellular senescence
Sandrine Morlot (IGBMC, Strasbourg)
NOTE THE NEW LOCATION (due to renovation work at LPS)
Saccharomyces cerevisiae is a powerful model organism to study replicative aging as asymmetric division gives rise to an aging mother cell and a rejuvenated daughter cell [Mortimer and Johnston 1959, Egilmez and Jazwinski 1989]. However the cellular mechanisms controlling replicative lifespan and the rejuvenation process are still poorly understood partly due to the technical limitations of following individual cells from birth to death. In this context, we have developed a high-throughput microfluidic device to follow up to 3200 single cells in parallel throughout their lifespan under the microscope. Thanks to this technology, we have established a timeline of events occurring successively during cellular aging. We have observed that cells experience a sharp transition into senescence. Indeed yeasts divide regularly every 90 minutes until a senescence entry point (SEP) which occurs after 20 generations. After this point, cell cycles strongly slow down until death [Fehrmann et al. Cell Reports 2013]. Furthermore we have measured that the SEP is preceded by an abrupt increase in the nuclear volume and more specifically in the size of nucleolus. The nucleolus is the nuclear compartment where ribosome biogenesis is initiated. This age-dependent nuclear defect is retained by the mother only, as daughter cells recover a normal nucleus and nucleolus, in agreement with the daughter cell rejuvenation paradigm. We have characterized that pre-ribosome particles accumulate in the nucleolus approximately 10 hours before entering into senescence. Our analysis suggests that this deficiency in ribosome biogenesis triggers cellular senescence.
Optimal immune systems
Aleksandra Walczak (LPT-ENS, Paris)
NOTE THE NEW LOCATION (due to renovation work at LPS)
Biological organisms have evolved a wide range of complex strategies to defend themselves against pathogens. I will present a common evolutionary framework that balances the benefits and costs involved in protection against pathogenic environments to maximize the long growth rate of populations. I will show that such a general evolutionary perspective recovers the basic forms of known immunity. I will then focus on adaptive immunity which is based on a combinatorically encoded repertoire of receptors that protects organisms from a diverse set of pathogens. A well-adapted repertoire should be tuned to the pathogenic environment to reduce the cost of infections. I will discuss a general approach for predicting the optimal repertoire that minimizes the cost of infections contracted from a given distribution of pathogens.
CTCF mediates allele-specific 3D domain structure at paternally imprinted gene loci
Daan Noordermeer (I2BC, Gif-sur-Yvette, France)
NOTE THE NEW LOCATION (due to renovation work at LPS)
Imprinted genes are mammalian genes where only one copy (allele) is active, depending on whether it is inherited from the mother or the father. This selective activity is determined by allele-specific DNA methylation at defined sites in the genome, so-called Imprinting Control Regions (ICRs). Recent microscopy studies by the group of Robert Feil (IGM-Montpellier, France) have revealed that imprinted genes are differently organized in the cell nucleus, depending on their parental origin (Kota et al., 2014). We have used high-resolution 4C-seq studies (Circular Chromosome Conformation Capture) to dissect the mechanisms and dynamics of this differential organization at the Dlk1-Dio3 and Igf2-H19 loci.
Recent studies have revealed that mammalian genomes are organized into Topologically Associating Domains (TADs) that demarcate ‘gene regulatory neighborhoods’ (Dixon et al., 2012). These physical domains are formed through a mechanism of ‘loop extrusion’ of the DNA fiber, with borders that are demarcated by opposing binding sites of the architectural CTCF protein (Fudenberg et al., 2016). I will show that the imprinted Dlk1-Dio3 and Igf2-H19 loci are organized into different DNA domains, determined by allele specific CTCF binding.
Both the Dlk1-Dio3 and Igf2-H19 loci are contained within large, invariant Topologically Associating Domains (TADs). The presence of the CTCF protein at the unmethylated ICRs on the maternal allele allows the establishment of new loops within the TADs. This result in the formation of a domain that acts like a cage, thereby shielding regulatory elements from nearby genes.
At the paternal alleles, DNA methylation at the ICR inhibits CTCF binding. As a result, the paternal Igf2-H19 allele displays little specific organization within the TAD. In contrast, at the paternal Dlk1-Dio3 locus, loops are formed between more distant unmethylated CTCF sites. The paternal allele therefore forms a much larger subdomain that is contained within the TAD.
Our work, for the first time, shows that constitutive TADs can have a markedly different allele-specific internal domain organization. Moreover, it shows that methylation-dependent DNA binding of the CTCF protein at ICRs guides the process of loop extrusion, thereby change the 3D structure of chromatin domains. We speculate that the imprinted patterns of gene expression at these loci are mostly imposed by the maternal 3D architecture, supporting previous genetic studies.
Tradeoffs between fast growth and adaptability shape microbial phenotypes
Markus Basan (ETH Zurich, Switzerland & Harvard University, USA)
SPECIAL LOCATION (due to renovation work at LPS)
Microorganisms exhibit a striking diversity of phenotypes in different conditions. Changes in growth rates are accompanied by large variations in metabolic strategies, gene expression and cell size. However, the molecular basis and underlying rationale of many of these complex patterns remains poorly understood. We illustrate how a quantitative approach, based on establishing empirical relations between cellular phenotypes, can help to elucidate such questions by focusing on three long-standing biological problems: the origin of overflow metabolism, the control of cell size and finally we provide an outlook on the emergence of severe, multi-hour lag phases. Coarse-grained models yield a quantitative and predictive understanding of phenotypic patterns under environmental as well as genetic perturbations and can even shed light on underlying molecular mechanisms. A common theme that emerges from these seemingly diverse questions is the existence of fundamental tradeoffs between fast growth and the ability to swiftly adapt to environmental changes or stress conditions.
Fluctuations in in vivo reactive systems.
Hélène Berthoumieux (LPTMC - Université Pierre & Marie Curie, Paris)
SPECIAL LOCATION (due to renovation work at LPS)
For a chemist, a living cell is a reactive system in which the concentrations of biomolecules are not determined by the thermodynamics but are controlled by energy sources maintaining the system in an out-of-equilibrium state. Chemical reaction networks are thus perturbed by a thermal noise and the fluctuations of these energy sources. Theoretical and experimental studies have shown that the fluctuations of in vivo systems break the fluctuation-dissipation theorem, which is a result of statistical physics at equilibrium. One can thus ask what information is contained in the correlation functions of protein concentrations and how they relate to the response of the reactive network to a perturbation. Answers to these questions are of prime importance to extract meaningful parameters from the in vivo fluorescence correlation spectroscopy data. Here, we present a theoretical study of the fluctuations of the concentration of a reactive species involved in a cyclic network that is in a non-equilibrium steady state perturbed by a noisy force, taking into account both the breaking of detailed balance and extrinsic noises that are known to be important in a cell.
Inferring interaction partners from protein sequences
Anne-Florence Bitbol (Laboratoire Jean Perrin, UPMC)
SPECIAL LOCATION (due to renovation work at LPS)
Specific protein-protein interactions play crucial roles in the stability of multi-protein complexes and in signal transduction. Thus, mapping these interactions is key to a systems-level understanding of cells. However, systematic experimental identification of protein interaction partners is still challenging, while a large and rapidly growing amount of sequence data is now available. Is it possible to identify which proteins interact just from their sequences? We propose an approach based on sequence covariation, building on methods used with success to predict the three-dimensional structures of proteins from sequences alone. Our method identifies specific interaction partners with high accuracy among the members of two ubiquitous prokaryotic protein families, and paves the way to identifying novel protein-protein interactions directly from sequence data.
Probing protein interactions on a microtubule bench by fluorescence microscopy: Application to YB-1, a mRNA-binding protein
David Pastré (Université d'Évry)
A typical procedure adopted by biologists to analyze protein interactions is to take advantage of the high throughput capability of the two hybrid system, generally in yeast, or that of the combination of affinity purification with mass spectrometry to obtain an exhaustive list of potential partners for a protein of interest. However, these assays require cell lysis, antibodies and adsorption onto non physiological substrates leading to false positives and false negatives. There is therefore a need to control the relevance of the proposed interactions in a context closer to native conditions, such as in living mammalian cells. To that end, novel methods are currently developed to provide a better view on protein interactions. In line with this, we have proposed a novel technology to detect and quantify direct or indirect protein interactions by fluorescence microscopy in living mammalian cells using microtubules as platforms. Microtubules are micrometer-long rigid cylinders of about 25 nm in diameter that are present in the cytoplasm of all eukaryotic cells. Due to their geometry, they provide an ideal surface to probe molecular interactions by fluorescence microscopy. As a proof of concept, we used microtubules to probe the interactions between mRNA-binding proteins like YB-1 in the cytoplasm.
High-Speed Atomic Force Microscopy: The dawn of dynamic structural biochemistry
Simon Scheuring (INSERM & Aix-Marseille Université)
The advent of high-speed atomic force microscopy (HS-AFM [1]) has opened a novel research field for the dynamic analysis of single bio-molecules: Molecular motor dynamics [2,3] membrane protein diffusion [4], assembly [5] and conformational changes [6] could be directly visualized. Further developments for buffer exchange [7] and temperature control [8] during HS-AFM operation provide breakthroughs towards the performance of dynamic structural biochemistry using HS-AFM.
[1] Ando et al., Chem Rev. 2014 Mar 26;114(6):3120-88.
[2] Kodera et al., Nature. 2010 Nov 4;468(7320):72-6.
[3] Uchihashi et al., Science. 2011 Aug 5;333(6043):755-8.
[4] Casuso et al., Nat Nanotechnol. 2012 Aug;7(8):525-9.
[5] Chiaruttini et al., Cell. 2015 Nov 5;163(4):866-79.
[6] Ruan et al., 2016, in preparation
[7] Miyagi et al., Nat Nanotechnol. 2016, in press
[8] Takahashi et al., 2016, in preparation
Active Composite Cell Surface - local control of clustering and sorting
Madan Rao (Raman Research Institute & National Centre for Biological Sciences-TIFR, Bangalore)
The surface of a living cell needs to regulate and control its local composition in a variety of contexts such as endocytosis and signalling. We have shown that many cell surface molecules are organised at multiple scales by their coupling to a thin cortical actomyosin layer, which actively drives local cell membrane composition and shape. This includes both transmembrane proteins and lipid anchored proteins, such as GPI-anchored proteins. Crucial to this engagement is the spontaneous emergence of localized contractile platforms in the 2dim active cortical fluid. Both the nanoscale clustering and the mesoscale segregation display the unique signatures of activity. We have recently recapitulated many of these effects in a minimal reconstituted system. I will end by discussing potential implications of this active composite cell surface for the processing of cellular information.
SEMINAR CANCELLED
Shaping a fly wing
Franck Jülicher (MPI-PKS Dresden)
A fundamental question in Biology is to understand the morphogenetic processes by which an organism of complex shape forms from a fertilized egg. This morphogenesis involves the dynamic remodeling of tissues consisting of many cells that grow and divide. The fly wing is an important model system for the study of multicellular dynamics during tissue morphogenesis. During pupal stages, the early fly wing undergoes a spectacular dynamic reorganization that involves cell flows, cell divisions and cell shape changes. This dynamic process generates the final shape of the wing. We characterize tissue remodeling by the contributions of specific cellular processes such as cell shape changes and cell neighbor exchanges to macroscopic shear at different times. We discuss the dynamics and the mechanics of this tissue using theoretical approaches that capture the essential physics of tissue remodeling. Our work suggests that local tissue contraction together with anisotropic active processes drive tissue remodeling in the fly wing. We show that mechanical boundary conditions play a key role in determining the final tissue shape.
Assessment of optimal parameters for deep optogenetic stimulations in non human primate
Frédéric Pain (IMNC, Université Paris-Sud)
SPECIAL LOCATION (next door from the usual one)
Optogenetics has become ubiquitous in fundamental neuroscience labs as a very powerful tool to unravel brain networks connectivity and cellular mechanisms. Yet, its clinical translation requires a careful assessment of the inocuity of repeated and sustained high power light stimulations. In a preliminary studies to translational research in the field deep brain stimulation for Parkinson's disease we have studied in vivo in anesthetized rats the potential damages and non-physiological effects produced by high power optical neurostimulation in typical optogenetics experiments.2D Maps of light distribution and temperature increase were recorded in wild type anesthetized rats brains for relevant optical stimulation protocols used in optogenetics. The spatial profile of light distribution and heat were correlated and demonstrated as expected a rapid attenuation with distance to the fiber. Temperature increase remains below physiological changes for stimulations up to 400mW/mm². I will present optogenetics issues in a clinical translational context.
Mechanics of B cell response
Paolo Pierobon (Institut Curie, Paris)
B lymphocytes are the antibodies producing cells and therefore essential effectors of adaptive immunity. In vivo, their activation is mostly triggered by the engagement of their B cell receptor (BCR) with antigens exposed at the surface of neighbouring antigen presenting cells. This leads to the formation of a signalling platform, the immune synapse, where cytoskeleton rearrangement are essential for the antigen extraction, internalization and processing. While it has been shown that on a hard substrate the cell follows a dynamics of spreading and contraction, this has never been investigated on substrates with rigidity close to the physiological one. We measure for the first time the forces produced by B cells on deformable antigen coated surfaces (traction force microscopy) and show that these forces are contractile, specifically induced by BCR activation and Myosin II dependent. We characterize the contractile dynamics of the cell and argue that in generating pulling forces, Myosin II plays a crucial role in antigen gathering and internalization. These results open interesting perspectives on the role of mechanics in the acquisition of specific antigen and more generally on receptor internalization.
Inferring anomalous diffusion from single particle trajectories
Denis Grebenkov (PMC, École polytechnique)
SPECIAL LOCATION
Transport of macromolecules, organelles and vesicles in living cells is a very complicated process that essentially determines and controls many biochemical reactions, growth and functioning of cells. The passive thermal diffusion through the overcrowded cytoplasm is combined with the active transport by motor proteins attached to microtubules. This intricate mechanism results in anomalous diffusions that found abundant experimental evidences but no consensus on the physical mechanism and the appropriate mathematical model is achieved so far. Single-particle tracking (SPT) experiments survey random trajectories of individual tracers inside living cells and can thus provide the missing information on the intracellular transport in order to discriminate between different physical mechanisms and to identify the appropriate theoretical model of anomalous diffusion. In SPT, an ensemble average of the quantities of interest (e.g., diffusivity, viscosity, first passage times, etc.) is often unavailable or even undesired, as tracers move in spatially heterogeneous and time evolving media such as living cells. One faces therefore a challenging problem of inferring dynamical, structural and functional properties of living cells from a limited (small) number of individual random realizations of an unknown stochastic process.
After a short introduction to theoretical aspects of the intracellular transport, we discuss the recent progress onto probing ergodicity of the tracer dynamics from a single particle trajectory. The proposed estimators are first investigated for several models of anomalous diffusion. In the case of nonergodic continuous time random walks, we show analytically that both estimators do not vanish even for infinitely long trajectories. The estimators are then applied to two sets of earlier published trajectories: mRNA molecules inside live E. coli cells and Kv2.1 potassium channels in the plasma membrane. These tests suggest that the former set exhibits ergodic behavior while the latter reveals both ergodic and nonergodic features.
Nanoparticules de type « cage » : applications pour lutter contre le cancer et les infections résistantes au traitement
Ruxandra Gref (ISMO, Université Paris-Sud)
Deux exemples de vecteurs de médicaments élaborés par une « chimie douce » (sans solvant) seront présentés : i) les nanoparticules hybrides organiques-inorganiques (metal-organic frameworks ou MOFs), qui sont des assemblages supramoléculaires cristallins hautement poreux, et ii) les nanoparticules à base de cyclodextrines, molécules « cage ». Avantageusement, ces nanoparticules sont capables d’encapsuler des quantités importantes de molécules thérapeutiques de nature chimique variée par simple imprégnation dans un milieu aqueux. La surface de ces vecteurs a été modifiée avec des éléments de reconnaissance spécifiques afin de moduler l’interaction des nanoparticules avec le milieu vivant et d’accroitre l’internalisation de celles-ci dans les cellules cible (cellules cancéreuses ou cellules infectées avec le VIH ou des bactéries). Finalement, des exemples seront présentés ou la libération des molécules thérapeutiques se fait « sur demande » suite à l’application d’un stimulus externe. Les applications de ces travaux se situent dans le domaine du traitement du cancer et des infections résistantes au traitement.
Tracking nonequilibrium physics in living systems
Étienne Fodor (Université Paris-Diderot)
Living systems operate far from equilibrium due to the continuous injection of energy provided by ATP supply. The dynamics of the intracellular components is driven by both thermal equilibrium fluctuations and active stochastic forces generated by the molecular motors. Tracer particles are injected in living cells to study these fluctuations. Alternatively, vesicles which are already present in the cytoplasm serve as probes of the intracellular dynamics.
To sort out genuine nonequilibrium fluctuations from purely thermal effects, we combine passive and active microrheology methods. They consist in measuring the spontaneous tracer fluctuations and extracting the response from an external oscillatory perturbation. By testing the fluctuation-dissipation theorem, we quantify the deviation from equilibrium appearing at low frequency. Removing the thermal contribution in the tracer fluctuations, we estimate the spectrum of the active forces. Eventually, we report non-Gaussian tails in the tracer displacement distribution as a result of directed motion events.
We recapitulate theoretically the observed fluctuations by modeling the dynamics with a confining harmonic potential which experiences random bursts as a result of motor activity [1]. This minimal model allows us to quantify the time and length scales of the active forces, along with the energy scale injected by the ensuing fluctuations [2, 3]. Finally, we estimate the energy dissipated by the tracers in the surrounding environment, leading us to define an efficiency of the energy conversion driving the tracer dynamics [4].
References- [1] É. Fodor et al., Phys. Rev. E 90, 042724 (2014)
- [2] É. Fodor et al., EPL 110, 48005 (2015)
- [3] W. W. Ahmed et al., arXiv:1510.08299
- [4] É. Fodor et al., arXiv:1511.00921
SEMINAR CANCELLED
Simulating Growing Tissues
Jens Elgeti (Forschungszentrum Jülich)
Growth of solid tumors or metastasis requires, besides massive biomedical changes, also a spatial remodelling of the tissue. This remodelling, often including displacements of healthy tissue around, requires mechanical work to be done. These mechanics of growth has attracted a lot of attention in recent years, but still remains poorly understood.
We use particle based simulations to study mechanical properties and effects in growing and motile tissues. These simulations have been helpful in understanding, interpreting and designing experiments. I will present an overview of the simulation technique, and how it contributed to recent developments in three dimensional tissue growth and collective cell migration. In a recent series of simulations and close experimental collaborations we found important interfacial and surface effects that lead to novel phenomena. For example, the tissue divides favorably at a free surface, even without any nutrient effects. This leads to the possibility and stability of a negative homeostatic pressure. In turn, a negative homeostatic pressure leads to naturally to finite steady states and tensile states.
References: [1] M.Basan et al, PNAS 110:2452 (2013) [2] F. Montel et al, N. J. Phys. 14:055008 (2012) [3] F. Montel et al, Phys. Rev. Lett. 107:188102, (2011) [4] M. Basan et al, Phys. Biol. 8:026014, (2011) [5] J. Ranft et al, PNAS 107:20863, (2010)
Insights on the regulatory principles of genome organization in unicellular microorganisms
Romain Koszul (Institut Pasteur)
Chromosomes of a broad range of kingdoms, from bacteria to mammals, are structured by large topological domains, whose precise functional roles and regulatory mechanisms remain elusive. Using chromosome conformation capture technology, we unraveled the higher-order organization of the Bacillus subtilis, Escherichia coli and Vibrio cholerae genomes, in a variety of growth and mutant conditions. Different types of topological domains were found to structure these chromosomes, ranging from a few dozens to a thousand kb. We show that the matP/matS and parB/parS systems generate specific types of topological structures, regulated by replication and cell cycle progression. We have also functionally characterized some of the global organizational principles of these domains, in link with replication/segregation during the cell cycle. Overall, the comparative analysis of these different species provide striking insights on the diversity of the regulatory mechanisms of genome structure of the bacterial world. In addition, I will also present and discuss recent data obtained during the cell cycle of the eukaryotic species Saccharomyces cerevisiae.
Regulation of actin assembly and mechanotransduction in cell-matrix adhesion complexes: a biochemical study of the talin-vinculin complex
Christophe Le Clainche (I2BC, Gif-sur-Yvette)
Cell migration is involved in many physiological and pathological processes. Force is produced by the growth and the contraction of the actin cytoskeleton (1). To produce force in adherent cells, these actin networks must be anchored to the extracellular matrix (ECM) by adhesion complexes (1,2). These structures contain transmembrane integrins that mechanically couple the ECM to the intracellular actin cytoskeleton via actin binding proteins (ABPs) (2). This system acts as a molecular clutch that controls force transmission across adhesion complexes. This molecular clutch is a complex interface made of multiple layers of regulated protein-protein interactions (2). The multiple activities of the ABPs present in these structures play a critical role in the dynamics of this interface. In addition to the control of actin filament binding and polymerization (1-3), these proteins sense and respond to the force applied by the actomyosin cytoskeleton to adjust the anchoring strength (4,5). Our goal is to determine the molecular mechanisms by which these ABPs cooperate to control the mechanical coupling between the actin cytoskeleton and cell-matrix adhesion complexes. To study these ABPs, our laboratory combines the measurement of actin polymerisation kinetics in fluorescence spectroscopy, single actin filament observations in TIRF microscopy and the reconstitution of actin-based mechanosensitive processes on micropatterned surfaces. Our model system is the mechanosensitive complex made of the two ABPs talin and vinculin. Our results showed that vinculin controls actin filament elongation (3). More recent results revealed that talin also regulates actin polymerisation in response to integrin binding (unpublished data). In addition, we have developed a microscopy assay with pure proteins in which the self-assembly of actomyosin cables controls the association of vinculin to a talin-micropatterned surface in a reversible manner (4, 5). This in vitro reconstitution revealed the mechanism by which a key mechanosensitive molecular switch senses and controls the connection between adhesion complexes and the actomyosin cytoskeleton. References- (1) Christophe Le Clainche and Marie-France Carlier. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiological Reviews (2008) Apr;88(2):489-513.
- (2) Corina Ciobanasu, Bruno Faivre, Christophe Le Clainche. Integrating actin dynamics, mechanotransduction and integrin activation: The multiple functions of actin binding proteins in focal adhesions. European Journal of Cell Biology (2013) (92) 339-348.
- (3) Christophe Le Clainche, Satya P Dwivedi, Dominique Didry, Marie-France Carlier. Vinculin is a dually regulated actin filament barbed-end capping and side-binding protein. Journal of Biological Chemistry (2010) Jul 23;285(30):23420-32.
- (4) Corina Ciobanasu, Bruno Faivre, Christophe Le Clainche. Actomyosin dependent formation of the mechanosensitive talin-vinculin complex reinforces actin anchoring. Nature Communications (2014) 5:3095
- (5) Corina Ciobanasu, Bruno Faivre, Christophe Le Clainche. Reconstituting actomyosin-dependent mechanosensitive protein complexes in vitro. Nature Protocols (2015) Jan ;10(1):75-89
Osmotic spreading of Bacillus subtilis biofilms
Agnese Seminara (LPMC, Université de Nice)
Bacterial biofilms are organized communities of cells living in association with surfaces. The hallmark of biofilm formation is a well defined spatio-temporal pattern of gene expression, leading to differentiation and a complex morphology. While this process resembles the development of a multicellular organism, biofilms are only transiently multicellular. More importantly the functions associated to the biofilm phenotype are largely unknown.
A common feature of biofilm formation is the secretion of a polymeric matrix rich in sugars and proteins in the extracellular space. In Bacillus subtilis, secretion of the exopolysaccharide (EPS) component of the extracellular matrix is genetically coupled to the inhibition of flagella-mediated motility. The onset of this switch results in slow expansion of the biofilm on a substrate. Different strains have radically different capabilities in surface colonization: Flagella-null strains spread at the same rate as wild type, while both are dramatically faster than EPS mutants. Multiple functions have been attributed to the EPS, but none of these provides a physical mechanism for generating spreading. We propose that the secretion of EPS drives surface motility by generating osmotic pressure gradients in the extracellular space. A simple mathematical model based on the physics of polymer solutions shows quantitative agreement with experimental measurements of biofilm growth, thickening, and spreading. We discuss the implications of this osmotically driven type of surface motility for nutrient uptake that may elucidate the reduced fitness of the matrix-deficient mutant strains.
Looking for the mechanical control of growth in plants. Is there a simple law?
Alexis Peaucelle (INRA Versailles/Cambridge University)
Plants are strikingly good at math, especially geometry. One could find parts or full plants shaped as spheres, circles, straight lines, and flat surfaces, golden and right angles and all sorts of exotic and pretty combination of shapes. These shapes are generated through complex tissue growth. We want to understand this puzzling beauty by focusing on the biophysical properties of the cell wall and its related biochemistry. We will present some of our results on dark grown hypocotyl and pavement cells demonstrating that pectin methylesterification status change is necessary for cell and tissue differentiation, growth and is related to changes in cell wall elasticity. Then we will expose puzzling results showing that cell growth rates are proportionate to the elastic stretching of the cell wall (Pressure divided by the Young modulus) and not plastic properties of the cell wall components. Finally, we will present preliminary experiments that could explain this paradox as well as some others such as microtubule partial correlation with oriented growth, and sound-induced plant growth.
Control of collective cell dynamics by adhesion, tension and fracking
Xavier Trepat (ICREA @ Institute for Bioengineering of Catalonia (IBEC), Barcelona)
A broad range of biological processes such as morphogenesis, tissue regeneration, and cancer invasion depend on the collective dynamics of epithelial cells. Such dynamics are determined by an exquisite balance between intercellular adhesion, cytoskeletal tension, and intracellular pressure. To study this balance in a fully quantitative manner I will present new techniques to map physical forces between and within cells. Using these techniques we studied how cellular forces are regulated and transmitted by the proteins that comprise intercellular adhesion complexes. To do so, we perturbed the main molecular players of the intercellular adhesome using RNAi and studied how these perturbations impact physical forces and cellular velocities in epithelial cell collectives. We found that perturbations targeting adherens junctions, but also tight junctions, gap junctions, and desmosomes have a significant impact on cell velocities, cell deformations, cell-matrix traction forces, and cell-cell forces. We developed a cross-validation analysis to show that concentrations of cell-cell adhesion proteins are significant predictors of cell-cell forces. Finally, I will discuss the determinants of epithelial integrity in the presence of stretching and transepithelial pressure.
Physics of active contractile matter
Ulrich S. Schwarz (Heidelberg University)
Biological systems such as cells and tissue use non-equilibrium processes to actively generate mechanical stress, movement and growth. Some of these processes can actually be reconstituted in biomimetic experiments with active soft matter. In this talk, we will first discuss why and how contractile forces are generated by biological systems and how they can be measured with soft elastic substrates ("traction force microscopy"). Because kilo-Pascal stresses are typically transmitted through micrometer-sized contacts, the relevant force scale is nano-Newton. We then introduce different theoretical approaches to understand and model the contractility of biological matter. Because closed systems have to conserve momentum, the most central concept here is the one of a "force dipole", similarly as for the theoretical description of microswimmers, but now coupled to a mechanical rather than to a hydrodynamic environment. We present a stochastic theory for the biologically most relevant example of a contractile force dipole, namely the "myosin II minifilament". We then explain why on the large length scale of cells and tissue, the mechanical properties of these systems are dominated by tensions rather than by their elastic modulus, with dramatic consequences for their shape and force transmission to the environment.
Growth of living fibrous tissues: from biofilms to fibrosis
Martine Ben Amar (ENS Paris)
Morphologies of soft materials in growth, swelling or drying have been extensively studied recently. Shape modifications occur as the size varies transforming ordinary spheres, cylinders and thin plates into more or less complex objects. Existence of fibers exacerbates this complexity, giving anisotropy to the growth process itself. The growth is coupled to the environment, for bacteria the substrate, in pathology the healthy tissue. In pathological situations such as wound-healing or desmoplastic tumor growth, the immune system reacts with a battery of morphogenetic gradients, making a new tissue full of collagene and eventually sending active cells. All these factors contribute to a high level of compressive stress at the origin of patterns and deformity. I will show how we can predict quantitatively these patterns on the simple drop geometry of the biofilms and on the spherical shape of tumors.
For the pathological cases, it turns out that the wrinkling process dominates the growth, deforming the tissues and exacerbating the immune system which reacts via passive (fibroblasts) and active cells (myo-fibroblasts). I will show that the consequence is a huge increase of the stiffness, which stops spontaneously when the healing is achieved but not in case of implants or tumors. Naive estimations can be given explaining difficulties encounted in drug treatments, for example.
Joint work with Min Wu.
Actomyosin Force Generation and Pattern Formation
Stephan Grill (MPI-CBG Dresden)
Morphogenesis is one of the great unknowns in Biology. Much is known about molecular mechanisms of regulation, but little is known about the physical mechanisms by which an unpatterned blob of cells develops into a fully structured and formed organism. The actomyosin cortex is a thin layer underneath the cellular membrane that can self contract, which drives many of the large-scale morphogenetic rearrangements that are observed during development. How this cortex reshapes and deforms, and how such morphogenetic processes couple to regulatory biochemical pathways is largely unclear. I will discuss two emergent physical activities of the actomyosin cytoskeleton, an active contractile tension and an active torque, both of which can serve to drive flows and large-scale chiral rotations of the actomyosin cytoskeleton. I will illustrate how active tension drive flows, how molecular constituents of the cortex affect flows, and how morphogenetic patterns can be formed by coupling regulatory biochemistry to active cortical mechanics. A particular focus will be the investigation of how compressive cortical flow drives the formation of an actin filament alignment pattern for generating a cleavage furrow for cytokinesis.The Dynamics of Two Biological Interfaces
Gerald G. Fuller (Stanford University)
Seminar co-hosted by Éric Raspaud—SPECIAL TIME
Biological systems are normally high-interface systems and these surfaces are laden with biological molecules and cells that render them mechanically complex. The resulting nonlinearities with response to surface stresses and strain are often essential to their proper function and these are explored using recently developed methods that reveal an intricate interplay between applied stress and dynamic response. Two applications are discussed.
1. Vascular endothelial cells are nature's "rheologists" and line the interior walls of our blood vessels and are sensitive to surface shear stresses. These stresses are known to affect the shape and orientation of endothelial cells. It is evident that the spatial homogeneity of flow can affect vascular health and it is well-documented that lesions form in regions of high curvature, bifurcations, and asperities in blood vessels. Experiments are described where stagnation point flows are used to create regions of well controlled flow stagnation and spatial variation of wall shear stresses. Live-cell imaging is used to monitor the fate of cells attached to surfaces experiencing flow impingement and it is revealed that endothelial cells migrate and orient in such flows to create remarkable patterns of orientation and cell densification. This response, termed "rheotaxis", is used to explore mechano-transduction pathways within these cells.
2. The tear film of the eye is a composite structure of an aqueous solution of protein and biomacromolecules. This thin layer is further covered by a film comprised of meibomian lipids excreted during each blink. The purpose of the meibum has been largely unexplained although one prevailing suggestion is that it suppresses evaporation. Recent measurements in our laboratory demonstrate that this layer is strongly viscoelastic and this property has dramatic effects on the dynamics of the moving contact line and stability against dewetting.
—
Gerald Fuller is the Fletcher Jones Professor of Chemical Engineering at Stanford University. He joined Stanford in 1980 following his graduate work at Caltech where he acquire his MS and PhD degrees. His undergraduate education was obtained at the University of Calgary, Canada. Professor Fuller's interests lie in studies of rheology and interfacial fluid mechanics. His work has been recognized by receipt of the Bingham Medal of The Society of Rheology, membership in the National Academy of Engineering, and honorary doctorates from the Universities of Crete, Greece, and Leuven, Belgium.
Stochasticity and robustness in growth and morphogenesis
Arezki Boudaoud (ENS Lyon)
How do organisms cope with natural variability to achieve well-defined morphologies and architectures? We addressed this question by combining experiments with live plants and analyses of stochastic models that integrate cell-cell communication and tissue mechanics. This led us to counterintuitive results on the role of noise in development, whereby noise is either filtered or enhanced according to the level at which it is acting.
The Cytoskeleton as an Active Gel: Modelling Cell Polarization, Shape Change, and Migration
Andrew Callan-Jones (Université Paris-Diderot)
Cell polarization and shape change are required for large-scale movements during embryo development and cancer metastasis. I will present recent work to understand these phenomena by studying two model systems: zebrafish embryos during gastrulation and confined HeLa cells. In both cases, individual cells are observed to undergo a novel type of polarization and transformation to a motile state that is crucially dependent on elevated levels of contractility in the actomyosin cortex. Polarization of zebrafish cells in vitro can be triggered by stimulating myosin activity: initially quasi-spherical, immobile cells switch to a polarized state characterized by a high cortical density at the cell rear, persistent cortical actin flows, and a distinctive pear-like morphology. Compressing HeLa cells between two plates results in a transition from a well-spread, mesenchymal-type migration mode to a rounded-up one sustained by cortical flow, and displaying an actin rich uropod at the rear, reminiscent of zebrafish. We have modeled these cell mechanical responses using active gel theory, a continuum-level description of out-of-equilibrium behavior of the cytoskeleton. In this talk, I will first provide a summary of this theory, and will then show how it accounts for the principal features of contractility-based polarization: cortical flow and density changes, cell shape change, and migration.How history shapes geometry in a model of protein evolution
Olivier Rivoire (Laboratoire interdisciplinaire de Physique, UJF Grenoble)
The interactions between amino acids in a protein are heterogeneous but not arbitrary: they enable proteins to perform specific biochemical "functions". Understanding these interactions may require, however, looking beyond current functional requirements, to the evolutionary history of proteins. I will illustrate this point with a simple statistical mechanics model, which I will motivate with observations and experiments on natural proteins. The model relates the parameters controlling the evolution of a protein to the organization of the interactions inside its structure.
Shape controlled filaments suspensions – rheology and dynamics
Thomas Gibaud (ENS Lyon)
The mechanical behavior of a suspension of rigid and semiflexible filaments has been studied in great detail. In comparison the effect of the filament geometry has been relatively unexplored. Here, we hijack flagellar filaments from their original purpose in order to develop a versatile model rod-like bio-colloid whose shape and length can be tuned. We present experimental results on the rheological behavior of suspensions of (1) straight, (2) curly and (3) semi-straight/semi-curly flagella with an identical average contour length. We find that (1) and (2) show an elastic behavior at intermediate time but that (3) remains elastic and does not flow at long times. Using fluorescence microscopy, we track individual filament and find that this elastic plateau is related to a cage in which the filament is trapped for a certain among of time. Taken together, this highlights the role of filament geometry in suspension mechanics.
Single cell analysis of entry into replicative senescence in budding yeast
Gilles Charvin (IGBMC Strasbourg)
Budding yeast cells have an asymmetrical division pattern. Each mother cell produces a limited number of smaller daughter cells before entering senescence and eventually dying. The detailed mechanisms that govern entry into senescence in mothers and daughter cell rejuvenation are still poorly understood. In this context, we have developed a microfluidic system that lets one monitor the successive divisions of single yeast cells in real-time under the microscope. Using this device, we have revisited classical paradigms associated with the age-dependent control of cell proliferation in this unicellular organism. Our results indicate that cells undergo a sharp transition to senescence, which is not related to the the loss of mitochondrial membrane potential, as previously proposed. Other applications of our methodology to the study of senescence induced by telomeres attrition and in other cellular biology contexts will be presented during the talk.
Hitching a ride at the nanometer scale: transport in passive and active complex media
Nikta Fakhri (Georg-August-Universität Göttingen)
Transport in crowded and complex media is a ubiquitous phenomenon in nature, which poses fundamental questions in statistical and soft matter physics. In particular, transport in the bustling interior of living cells is fascinating and far from understood. On a molecular scale, transport can be diffusive or driven, either externally or by local force generators. In this talk, I will introduce single-walled carbon nanotubes (SWNTs) as highly versatile multi-scale probes to investigate different modes of transport in media of increasing complexity: from the confined dynamics of semiflexible polymers in crowded environments to random stirring generated by non-equilibrium dynamics of the cell cytoskeleton.
Mechanics and growth of the actin cytoskeleton probed by magnetic micro-objects
Julien Heuvingh (PMMH, ESPCI)
The ability of cells to perform essential processes such as migration or deformation relies on their cytoskeleton, and especially on the structures and networks formed by the actin polymer and its associated proteins. Understanding the dynamics and the mechanics of the actin filaments and its multiple partner is a major goal at the frontier of biology and physics. Our team developed a new experimental setup to study the mechanics of in vitro reconstituted actin networks, with an unprecedented throughput. This technique is based on self-organized chains of micron-size magnetic beads or cylinders where the controlled attractive dipolar force between the colloids deforms the actin networks. We characterized for the first time the mechanics of actin networks reconstituted with different concentrations of purified proteins, leading to networks of different architectures, and drew conclusions on the origin of the elasticity on these networks (Pujol et al PNAS 2012). We are now measuring mechanical properties of networks reconstituted from yeast extract which allows comparison between a wild type containing hundred different actin binding proteins to mutants lacking some of them. Our experimental setup was decisively improved by the fabrication of magnetic micro-objects of cylindrical or cubic shape (Tavacoli et al. Soft Matter 2013) allowing the deformation of actin networks between two flat surfaces. In this way, we can access properties of dense branched networks such as non-linear elasticity and Poisson modulus, which are required to test theoretical models of fiber networks (Mikado). We are currently studying the growth velocity of the actin gel as a function of an applied mechanical stress and the architecture of the networks. I will also present other applications of our magnetic methods to probe the mechanics of whole cells.
[caption id="attachment_26491" align="aligncenter" width="502"] Actin networks (green) growing from the side of magnetic cylinders. Superimposition of bright field image (gray) and fluorescent image (green). Cylinder length is ~12µm.[/caption]
Actin networks (green) growing from the side of magnetic cylinders. Superimposition of bright field image (gray) and fluorescent image (green). Cylinder length is ~12µm.[/caption]Understanding the self-assembly of simple icosahedral viruses
Guillaume Tresset (Université Paris-Sud)
Viruses are ubiquitous pathogens in all kingdoms of life and are major public health issues as well as economic and veterinary concerns worldwide. Despite a huge body of work dedicated to the molecular biology of viral life cycles, there are currently no physical models accounting reliably for the mechanisms by which the hundreds of molecular building blocks making up a virus fit into the final structure with a pinpoint accuracy. I will first present the self-assembly pathway of empty icosahedral capsids derived from a bovine virus. A kinetic model constructed from time-resolved X-ray scattering data reveals a cooperative mechanism involving an unexpected long-lived intermediate species. Then, I will give some insights into the packaging of polyelectrolytes by capsid proteins derived from a plant virus. Accurate measurements of the mass of packaged polyelectrolyte demonstrate a nonspecific selectivity that may play a crucial role for genome packaging in host cells. Quite generally, physics provides a useful framework to describe viral self-assembly and should eventually support the development of novel therapeutic strategies.
Bioimaging and quantum sensing with ion-irradiated nanodiamonds
Huan-Cheng Chang (Academia Sinica, Taiwan)
Seminar co-hosted by François Treussart SPECIAL TIME AND LOCATIONAs a wide band-gap material, diamond can contain a variety of atomic defects or impurities as color centers. Some of the color centers are highly luminescent, while others are luminescent with a very low quantum yield. For nanoscale diamonds (NDs) containing a high-density ensemble of vacancy-related defect centers, they are useful as nanoprobes for bioimaging and quantum sensing both in vitro and in vivo. In this seminar, we will show how ion-irradiated NDs can be routinely produced in our laboratory. Three examples of the applications by utilizing nitrogen-vacancy (NV−) centers and neutral vacancy (V0 or GR1) centers in NDs are discussed. First, we will present our results of using fluorescence lifetime imaging microscopy to achieve background-free real-time imaging of fluorescent NDs (denoted as FNDs) in living organisms such as C. elegans. With 100-nm FNDs conjugated with yolk lipoprotein complexes, we demonstrate that the nanoparticles serve well as a biomolecular nanocarrier without significantly altering the functionality of the cargos for intercellular transport, cell-specific targeting, and long-term imaging applications in vivo. Second, we report our recent work on the development of highly ion-irradiated NDs (denoted as INDs) as a photoacoustic contrast agent for deep-tissue imaging. The particles are so extensively damaged that graphitization occurs concurrently with the generation of the GR1 centers. Although the IND of ~40 nm in diameter has a much smaller absorption coefficient than gold nanorods (GNRs) of similar dimensions at 1064 nm, it shows a better performance due to higher thermal stability and a lower nanobubble formation threshold of the carbon-based nanomaterial. Finally, we apply the NV− centers in 100-nm FNDs for nanoscale temperature sensing by optically detected magnetic resonance. We conjugate FNDs with GNRs and employ them as both a nanoheater and a nanothermometer in solution and cells. The integration of heating and temperature sensing functions on the same particles opens an opportunity for active and high-precision control of temperature at the nanoscale by pure optical means.
Relaxation in Cell Cytoskeleton
Manuel Théry (Hôpital Saint-Louis, Paris)
Using combinations of in vivo and in vitro approaches we try to unravel the mechanisms regulating actin bundle and microtubule mechanical properties in response to geometrical and/or mechanical stimulations.Diffusion-controlled reactions in complex media
Francesco Piazza (Université d'Orléans)
In all biochemical reactions occurring in living tissues, reactants have to form an encounter complex before the specific chemical step. Invariably, in order to reach their binding partners, biomolecules have to diffuse in complex environments, both very crowded with all sorts of other biomolecules and organelles and confining, due to the presence of different membranes and cytoskeletal structures that strongly compartimentalize the available space.
Under such conditions, the standard Smoluchowski theory for biomolecular encounters valid in ideal solutions is no longer applicable and the need emerges for more sophisticated theoretical paradigms accounting explcitly for crowding and confinement in the computation of encounter rates.
In this talk, I will illustrate a general theoretical paradigm that we are developing in our group to solve this problem. Using addition theorems for spherical harmonics, we compute the diffusion rate to a sink in the presence of crowding agents that we model as spheres of arbitrary radius and endowed with arbitrary reactivity, from fully reflecting (purely excluded volume) to fully absorbing (competitive binding partners). We consider both diffusion in an unbounbded domain and diffusion occurring within a spherical domain, as an attempt to model encounters occurring within a cell. Different applications will be discussed, such as diffusion to a binding pocket in a coarse-grained model of protein and reactions occurring in vesicles and other kinds of nanoreactors.
SEMINAR CANCELLED
Leukocyte sensing of flow direction
Marie-Pierre Valignat (Laboratoire Adhésion Cellulaire et Inflammation, Marseille)
As they leave the blood stream and travel to lymph nodes or sites of inflammation, leukocytes are captured by the endothelium and migrate along the vascular wall to permissive sites of transmigration. These processes are supposedly orchestrated by chemical signals and take place under the influence of a strong hemodynamic shear stress. The role of flow on leukocyte crawling and extravasation remains generally an unsolved question, however crawling T lymphocytes were recently reported in vivo and in vitro to orient against the direction of flow and to move upstream like salmons in a river. This non-intuitive behavior is manifestly not a passive drift of cells pushed by the flow, and we sought here to clarify the origin, role and mechanism of this upstream flow mechanotaxis behavior.
Living soft matter
Gijsje Koenderink (FOM Institute AMOLF)
One of the defining qualities of soft matter is that it is readily driven far from thermodynamic equilibrium by external stress. Driving forces such as those due to an electric field or shear can drive colloidal suspensions and polymer networks into fascinating non-equilibrium patterns, such as banded or ordered steady states. By contrast, living cells naturally exhibit a unique form of internal driving in the form of chemomechanical activity. A prominent example is the cytoskeleton, a meshwork of protein polymers and force-generating motor proteins that constitutes the scaffold of cells. The cytoskeleton is responsible for driving vital cellular functions such as growth, division, and movement. In this talk, I will present two examples of our research on active cytoskeletal polymer gels. The first example concerns active contractility of the actin cortex, which lies underneath the cell membrane and drives shape changes by means of myosin motors. By reconstituting a simple model system composed of purified proteins, we could show how myosin motors and actin filaments collectively self-organize into force-generating arrays. We discovered that motors contract actin networks only above a sharp threshold in crosslink density, corresponding to a connectivity percolation transition. Surprisingly, the motors tend to drive initially well-connected networks robustly to this critical point. The second example I will discuss concerns cell shape polarization directed by interactions of actin filaments with microtubules. I will show that active force generation by growing and shrinking microtubules leads to feedback between the organization of the actin filaments and microtubules, explaining earlier observations made in living cells.
Navigating the cytoskeleton: novel tools to dissect and direct intracellular transport
Lukas Kapitein (Universiteit Utrecht, The Netherlands)
Active transport is important for proper cellular organization and functioning. Such transport is driven by a large variety of molecular motor proteins that can walk over cytoskeletal biopolymers such as microtubules and F-actin. Whereas controlled biophysical experiments using purified components have revealed many of the basic properties of these fascinating machines, much less is known about their specific intracellular activity and about the interplay between cytoskeletal organization and transport. To address these questions, we have developed novel tools to control the activity of specific motors inside cells. These experiments have revealed different mechanisms by which the underlying organization of the microtubule network guides motor transport to specific destinations. In addition, these tools enabled us to remote-control intracellular transport and alter cellular behavior using light.
Lukas Kapitein is assistant professor at the Division of Cell Biology of Utrecht University, where his group develops novel approaches to understand how the cytoskeleton and their associated motor proteins contribute to cellular organization and morphology. The combined use of well-controlled, inducible intracellular transport assays and fluorescence nanoscopy of the cytoskeleton offers unique insights into the interplay between cytoskeletal organization and motor-driven transport.
Etude multi-échelle des tissus riches en collagène
Jean-Marc Allain (École Polytechnique)
Nous nous intéressons au lien entre l'organisation du collagène dans les tissus mous (comme la peau ou les tendons) et leurs propriétés mécaniques. Pour cela, nous avons mis au point avec le LOB de l'Ecole Polytechnique un montage original qui combine une machine de traction avec un microscope à Génération de Seconde Harmonique. Cette microscopie non-linéaire permet d'image les fibrilles de collagène en 3D et sans marquage dans un tissu, donnant accès à son organisation à l'échelle micrométrique. We avons validé ce dispositif sur le tendon, avant de l'utiliser sur d'autres tissus.
Elasticity and wrinkled morphology of Bacillus subtilis pellicles
Éric Raspaud (LPS Orsay)
Bacterial biofilms refer to communities of bacteria that self-assemble into an extracellular cohesive matrix on a surface. We are recently interested in floating biofilms formed by wild strains of Bacillus subtilis on liquid medium. Wrinkles appear during their maturation. We have studied the formation of wrinkles in relation to their mechanical property and have shown that they could be due to a buckling instability. In this talk I will present our experimental results and their theoretical interpretations.
[caption id="attachment_22671" align="aligncenter" width="300"] Top view of bacterial pellicles floating on liquid media. Two wild strains of Bacillus subtilis are shown in Figure A and B.[/caption]
Top view of bacterial pellicles floating on liquid media. Two wild strains of Bacillus subtilis are shown in Figure A and B.[/caption]
Trejo M., C. Douarche, V. Bailleux, C. Poulard, S. Mariot, C. Regeard, E. Raspaud, Elasticity and wrinkled morphology of Bacillus subtilis pellicles. Proc Natl Acad Sci USA 110 (2013), 2011-2016.
Role of membrane elasticity in clathrin-mediated endocytosis
Aurélien Roux (Université de Genève, Suisse)
Aurélien Roux (Université de Genève, Suisse)
In Clathrin-mediated endocytosis, Clathrin assembles into a soccerball-like structure at the plasma membrane that was proposed to deform the membrane by scaffolding. However, controversies in the community have appeared on the exact role of Clathrin: does its polymerization force is sufficient to curve the membrane, or deformation by other means (protein insertion) is required? We studied the formation of Clathrin buds from Giant Unilamellar Vesicles, and found that the pits can be flattened when membrane tension is increased. This suggested that the Clathrin polymerization force could be counteracted by membrane tension, which we further proved by directly measuring Clathrin polymerization force: by pulling a membrane tube out of a GUV aspirated in a micropipette, we can measure the force required to hold the tube through an optical tweezer system. When Clathrin is added, it polymerizes onto the GUV predominantly, and the force drops. From these measurements, we can deduce that the polymerization strength of Clathrin is in the range of a few hundred micronewtons per meter. This value confirms that clathrin polymerization can be counteracted efficiently by membrane tension. To finalize endocytosis, the clathrin-bud needs to be separated from the plasma membrane. Membrane fission requires the constriction and breakage of a transient neck, splitting one membrane compartment into two. The GTPase Dynamin forms a helical coat that constricts membrane necks of Clathrin-coated pits to promote their fission. Dynamin constriction is necessary but not sufficient, questioning the minimal requirements for fission. Here we show that fission occurs at the edge of the Dynamin coat, where it is connected to the uncoated membrane. At this location, the specific shape of the membrane increases locally its elastic energy, facilitating fission by reducing its energy barrier. We predict that fission kinetics should depend on tension, bending rigidity and the Dynamin constriction torque. We verify that fission times depend on membrane tension in controlled conditions in vitro and in Clathrin-mediated endocytosis in vivo. By numerically estimating the energy barrier from the increased elastic energy, and measuring the Dynamin torque, we show that: 1- Dynamin torque, ≈1nN.nm, is huge but necessary to achieve constriction, and 2- Dynamin work sufficiently reduces the energy barrier to promote spontaneous fission.
Inhibitory signalling to the Arp2/3 complex steers cell migration
Alexis Gautreau (LEBS - Gif-sur-Yvette)
Cell migration requires the generation of branched actin networks that power the protrusion of the plasma membrane in lamellipodia. The Arp2/3 complex is the molecular machine that nucleates these branched actin networks. This machine is activated at the leading edge of migrating cells by the WAVE complex. The WAVE complex is itself directly activated by the small GTPase Rac, which induces lamellipodia. However, how cells regulate the directionality of migration is poorly understood. Here we identify a novel protein that inhibits the Arp2/3 complex in vitro, Arpin, and show that Rac signalling recruits and activates Arpin at the lamellipodial tip, like WAVE. Consistently, upon depletion of the inhibitory Arpin, lamellipodia protrude faster and cells migrate faster. A major role of this inhibitory circuit, however, is to control directional persistence of migration. Indeed, Arpin depletion in both mammalian cells and Dictyostelium discoideum amoeba resulted in straighter trajectories, whereas Arpin microinjection in fish keratocytes, one of the most persistent systems of cell migration, induced these cells to turn. The coexistence of the Rac-Arpin-Arp2/3 inhibitory circuit with the Rac-WAVE-Arp2/3 activatory circuit can account for this conserved role of Arpin in steering cell migration. Loss of this inhibitory circuit promotes exploratory behaviors and might commit carcinoma cells to the invasive state.
Fluorescent Nano-objects For Bioimaging Applications
Yang Si (ENS Cachan)
Special seminar: poster prize from the NOMBA workshop
Bacteria are the most abundant organisms in the world. Studying models of bacterial chromosome dynamics in the cytoplasm is very important to understand how bacteria adapt to different growth environments and in response to stimuli. Optical labeling is one of the most common methodologies used for bioanalytical purposes. The fundamental issues for any fluorescent material are the same: brightness and stability. In the quest for very bright and stable labels, novel polymer-based, self-stabilized, fluorescent nanoparticles (FNPs) and fluorescent polymer chains (FPCs) have been developed in the PPSM laboratory. They are brighter, more stable, photobleach slowly and are more easily functionalized compared with other fluorescent labels like GFP and QDs. A methodology to insert these FNPs (60nm) into E.coli bacteria was developed. To control if the FNP are indeed internalized, we developed a protocol based upon FNP luminescence quenching by methylene blue. Biotin conjugated FNPs could be used to study specific membrane proteins. By using a strepdavidin-biotin link, we made a “Sandwich” to build a bridge between particles, specific antibodies and bacteria. Negatively charged FPCs can easily enter into E.coli bacteria. It is found that FPCs can label the cytoplasm but not the DNA, which appears to be more compact. These unique properties will allow the study of DNA and cytoplasm viscosity changes during bacterial growth.
Optimal Design of Elongating Yeast Spindles
François Nédélec (EMBL Heidelberg)
Joint seminar with LEBS Gif-sur-Yvette
SPECIAL TIME AND LOCATION
Bundles of filaments are universal elements that belong to the intracellular skeleton of eukaryotic cells. The anaphase spindle from fission yeast is an excellent example of such bundle that assembles repeatedly in the same manner at every cell division. It is also an excellent experimental system since yeast cells can easily be perturbed genetically, and their spindle is visualized using electron or light microscopy. The spindle is under compression at anaphase and based on electron tomographic reconstructions of its constituent microtubules, we calculated that the length and organization of microtubules within the fission yeast spindle are optimized to achieve maximal strength while minimizing the use of material. A combination of simulations and live cell imaging further indicated which of the properties of the microtubule cross-linkers are likely to be responsible for such a precise regulation of spindle morphology, and the synergy that exists between the cross-linkers in fission yeast.
The hair-cell bundle as a mechanosensor and amplifier for hearing
Pascal Martin (Institut Curie - Paris)
The ear works as a remarkable sound detector. Hearing can indeed operate over six orders of magnitudes of sound-pressure levels, with exquisite sensitivity and sharp frequency selectivity to weak sound stimuli. Curiously, the ear does not work as a high-fidelity sound receiver, introducing in the auditory percept “phantom” tones that are not present in the sound input. In this talk, I will present micromechanical experiments at the level of the cellular microphone of the inner ear – the hair cell – whose function is to transduce sound-evoked vibrations into electrical nervous signals. In particular, I will show that hair cells can power spontaneous oscillations of their mechanoreceptive hair bundles, a tuft of cylindrical protrusions that protrudes from the apical surface of each cell. The oscillatory instability is thought to result from a dynamical interplay between ion channels, elastic proteinous linkages and active molecular motors. We find that oscillations of the hair bundle allow the hair cell to actively resonate with its mechanical input at the expense of distortions with properties that are characteristic of hearing. I will conclude by arguing that our results promote a general principle of sound detection that is based on nonlinear amplification by self-sustained “critical” oscillators in the inner ear, i.e. active dynamical systems that operate on the brink of a Hopf bifurcation.

Conformation and dynamics of DNA in confined environments: cross-talk between chromosomes in the nucleus and polymers in nanochannels
Aurélien Bancaud (Laboratoire d'analyse et d'architecture des systèmes, Toulouse)
Genome structure and dynamics attacts considerable attention in the biology community to eludicate genome regulation principles, but also for biological physicists who aim to develop models of DNA in vivo. The challenges of this research is conceptual but also economical because of expected impact of DNA sequencing or DNA microarrays technologies in personalized diagnostics.
Our research is carried out at the nexus of technology and biology and aims to provide a physical description of the genome structural properties. We will first overview our results on chromosome dynamics in living yeast, showing the unexpected flexibility of these structures in vivo. We will then focus on new methods for chromosome analysis in vitro based on micro- and nano-fluidics, and we will finally emphasize that that these two topics are not so unrelated, given that the physics of DNA confined environment can be used as a common research framework.
Looking at transcription mechanisms with single molecule FRET
Emmanuel Margeat (Centre de Biochimie Structurale, Montpellier)
Exceptional seminar hosted by Karen Perronet
SPECIAL DATE, TIME AND LOCATION
Förster Resonance Energy Transfer (FRET) allows measuring the distance between two spectrally distinct fluorophores, in the 20-100 Å range. When monitored at the single molecule level, smFRET is useful in resolving subpopulations, or observing conformational changes as a function of time within single macromolecular complexes in vitro. I will describe here the methodologies used to measure FRET accurately on single biomolecular complexes, freely diffusing in solution or immobilized on surfaces. I will then focus on our studies on the mechanism of prokaryotic transcription such as antitermination control by the antiterminator LicT, and Rho-induced transcription termination.
Role of mechanical constraints on the establishment of neocortical organisation
Roberto Toro (Institut Pasteur)
The mammalian brain is astonishingly diverse. Not only its size varies several orders of magnitude – from the 3 grams of the mouse brain to the 6 kg of the blue whale brain – but also its geometry and function. There is indeed a striking, largely unexplained, relationship between the folding of the mammalian brain and its cellular, functional and connective organisation. Brain folding appears as much more than a mere mechanical epiphenomenon, and besides its major evolutionary relevance, many psychiatric disorders such as autism or schizophrenia, are related to changes in brain folding.
I will present a brief overview of the developmental processes leading to the folding of the brain, and show some examples of functional correlates of brain folding in humans and other mammalian species. Finally, I will discuss some of the current theories proposed to explain the mechanism underlying the relationship between brain geometry and brain organisation, including our ongoing project on computational modelling and analysis of the development of the ferret brain.
Molecule motion inside secretory granules before and during exocytosis
Daniel Axelrod (University of Michigan)
SPECIAL DATE & TIME
Despite ~10% of the human genome being comprised of secretory proteins, little is known about the dynamics of proteins inside the secretory granule before and during fusion with the plasma membrane. This talk presents early results on measuring the diffusion coefficient of two proteins within that submicroscopic and closed space, and relates the results to secretion and local membrane deformation rates during the fusion event. The techniques used, TIR-FRAP, TIR-FCS, and pol-TIRF have general applicability, and the theory and practice of especially the first two is discussed.
The mechanics of active and passive cellular assemblies: How biomimetic reconstitution can help to understand living cell
Timo Betz (Institut Curie - Paris)
Understanding the intriguing complexity of living systems is one of the main driving forces of science. To gain insight we use biomimetic systems that reconstitute well defined cellular assemblies and compare these to the living system. Our main interests are the mechanical properties and the generation of forces, both mediated by the cytoskeleton and its interaction with the plasma membrane. Recent advances allow to mimic structures such as the actin cortex, sparse actin networks and actin bundles, and we use optical tweezers to quantify the mechanical properties of these structures and to compare them to living cells. While sparse actin networks and polymerizing actin bundles show rather passive behavior, we apply the same measurement methods to living cells such as cell blebs and red blood cells which allow to study the out-of-equilibrium mechanics of these systems, and to determine the timescale at which the system's activity becomes evident.
Division Control in Escherichia coli is Based on a Size-sensing rather than Timing Mechanism
Marie Doumic-Jauffret (INRIA Rocquencourt)
Models describing the growth of cell populations have been developed based on assumptions on the stochastic mechanisms underlying growth and division at the single cell level. In particular, two different models have been widely used for decades, assuming that cell division probability depends respectively on cell age (the renewal equation) or cell size (the size-structured or growth-fragmentation equation) - or both.
We confront these models with data on E. coli single cells growth, and develop a new estimation methodology, based on nonparametric functional testing within the PDE models, in order to test the hypothesis of an age-dependent versus size-dependent division rate. We conclude that in E. Coli, the division is controlled by a size-sensing rather than timing mechanism.
This is a joint work with L. Robert and M. Hoffmann.
Muscle power-stroke as a collective mechanical phenomenon
Matthieu Caruel (Inria and École Polytechnique)
The mechanism of muscle contraction, residing in nano-scale interaction between actin and myosin filaments, was intensely studied by using fast loading protocols. These experiments revealed the important mechanical role of the internal conformational change inside protruding myosin heads known as the power-stroke. It was realized that fast force recovery after abrupt loading, taking place at 1 ms time scale, is a purely mechanical phenomenon linked exclusively to the power-stroke and not limited by metabolic fuel delivery.
In this presentation, we explore previously unnoticed difference in fast force recovery taking place in hard and soft loading devices and propose a purely mechanical model that explains the origin of this unusual behavior. We link the inequivalence of soft and hard loading ensembles to the presence of long range interactions between the individual actin-myosin cross-links known as cross-bridges. Our fit of experimental data suggests that 'muscle material' is finely tuned to perform close to a critical point which explains large fluctuations observed at stall forces. The proposed mean field model clarifies the collective nature of the power-stroke and reveal new properties of the celebrated Huxley and Simmons 1971 model.
Fluorescence Microscopy of Biostructures @ Molecular Optical Resolution
Christoph Cremer (Institute of Molecular Biology (IMB), Mainz; Kirchhoff-Institute for Physics (KIP) and Institute for Pharmacy and Molecular Biotechnology (IPMB), University Heidelberg )
SPECIAL DATE & TIME
Seminar hosted by Olivier Acher & Guillaume Dupuis
Conventional epifluorescence microscopy is limited in resolution (to about 200 nm laterally, 600 nm axially) by the shere nature of light (by diffraction), and is hence insufficient to study the nanostructure of subcellular components.
At IMB-Mainz and Heidelberg University we have established a variety of superresolution microscopy ("nanoscopy") methods, for example Structured Illumination SMI and Localisation microscopy SPDMphmyd with blinking dyes, like standard GFP. Our microscope systems can and have been applied to study the composition, function and metabolism of many biomolecular structures and small paticles like single viruses in high densities. Currently we reach a resolution down to 5 nm in 2D and 40 nm in 3D in the co-localization mode.
There are various applications in the fields of molecular biology, (clinical) medicine, diagnosis and pathology.
- Christoph Cremer, Barry R. Masters (2013) Resolution enhancement techniques in microscopy, The European Physical Journal H 38, 3, pp 281-344
- Kaufmann R, Muller P, Hausmann M and Cremer C (2011). Imaging label-free intracellular structures by localisation microscopy. Micron, 42, 348-352.
- Kaufmann R, Müller P, Hildenbrand G, Hausmann M and Cremer C (2011) Analysis of Her2/neu membrane protein clusters in different types of breast cancer cells using localization microscopy. Journal of Microscopy, 242, 46-54.
- Gunkel M, Erdel F, Rippe K, Lemmer P, Kaufmann R, Hoermann C, Amberger R and Cremer C (2009). Dual color localization microscopy of cellular nanostructures. Biotechnology Journal, 4, 927-938.
Mechanotransduction in Vascular Endothelial Cells: Mechanisms and Implications
Abdul Barakat (École Polytechnique)
The ability of arterial endothelial cells, the cells lining the inner surfaces of blood vessels, to respond to mechanical forces associated with blood flow is essential for normal vascular function. Abnormalities in endothelial cell mechanotransduction play a critical role in the development and progression of cardiovascular disease. The mechanisms governing how endothelial cells sense mechanical forces on their surfaces and how they subsequently transmit these forces within the intracellular space remain poorly understood. In this talk, I will present experimental and computational results in support of a central role for the cellular cytoskeleton in force transmission within endothelial cells. Because endothelial cells are often simultaneously exposed to multiple biophysical stimuli, I will show data that demonstrate that endothelial cells integrate biophysical stimuli derived from simultaneous apical cellular stimulation by flow and basal stimulation by nano-scale substrate patterning. Finally, I will discuss the role of proteins that link the cytoskeleton to the nucleus in modulating mechanotransduction in endothelial cells.
Mechanical Force Generation and Turnover in the Cell Cytoskeleton
Michael Murrell (University of Wisconsin, Madison)
Myosin II motors drive contractility of the cortical actin network, enabling shape change and cytoplasmic flows underlying diverse physiological processes ranging from cell division and migration to tissue morphogenesis. Yet, despite its importance, the mechanisms that describe contractility and the generation of mechanical forces within the cortex are not well understood. We recapitulate contractility in vitro, through the development of a minimal model of the cell actomyosin cortex by coupling a two-dimensional, cross-linked F-actin network decorated by myosin thick filaments to a model cell membrane. Myosin motors generate both compressive and tensile stresses on F-actin and consequently, induce large bending fluctuations. Over a large range of crosslinking, we show the extent of network contraction corresponds exactly to the extent of individual F-actin shortening via buckling. This demonstrates an essential role of buckling in facilitating local compression to enable mesoscale network contraction of up to 80% strain. Buckled F-actin at high curvatures are prone to severing and thus, compressive stresses mechanically coordinate contractility with F-actin severing, the initial step of F-actin turnover. Finally, the F-actin curvature acquired by myosin-induced stresses can be further constrained by adhesion of the network to a membrane, accelerating filament severing but inhibiting the long-range transmission of the stresses necessary for network contractility. Thus, the extent of membrane adhesion can regulate the coupling between network contraction and F-actin severing. These data demonstrate the essential role of the non-linear response of Factin to compressive stresses in potentiating both myosin-mediated contractility and filament dynamics.
Dynamics of transcription and error incorporation in a viral RNA-dependent RNA polymerase
David Dulin (Delft University of Technology)
RNA-dependent RNA polymerases (RdRPs) are essential enzymes that govern transcription and replication in RNA viruses. While RNA elongation forms an important therapeutic target against viral infection, little is known about elongation dynamics at the single-molecule level. Here, we study the well-established RdRP model system of P2 from the double-stranded RNA bacteriophage Φ6 using high-throughput single-molecule force-spectroscopy combined with theoretical modeling. We show that P2 elongation dynamics is irregular, with rapid transcription repeatedly interrupted by pauses whose durations vary from seconds to thousands of seconds. Exploiting the discriminatory power offered by our large datasets, together with specifically-adapted analysis, we introduce a stochastic dynamical model of P2 transcriptional elongation. Our results imply that the majority of pauses result from nucleotide misincorporation, providing a direct link between RdRP dynamics and error rates—rates that offer potential drug targets, as they must be finely tuned to confer both genome stability between generations and adaptability to bypass host defense systems.
Magnetic living cells: New tools for cell imaging, tissue engineering and cell therapies
Claire Wilhelm (Université Paris 7)
Recent advances in cell therapy and tissue engineering opened new windows for regenerative medicine, but still necessitate innovative techniques to create and image functional tissues. One promising approach is to associate magnetic nanoparticles with cells in order to supply them with sufficient magnetization to be detectable by MRI or manipulated by magnetic forces, while maintaining cell viability and functionalities. A few years ago, we proposed the use of anionic iron oxide nanoparticles as efficient agents for cell internalisation without impacting cell functions. Recently we examined the influence of the amount of internalized iron and the state of nanoparticle aggregation on the capacity for mesenchymal stem cell differentiation and MRI single cell tracking. We then demonstrated that high resolution Magnetic Resonance Imaging (MRI) allowed combining cellular-scale resolution with the ability to detect two cell types simultaneously at any tissue depth. In parallel, we addressed the challenge to create a functional tissue from stem cells in vitro. The aim was to confine stem cells in three dimensions at the millimetric scale by using home-designed miniaturized magnetic devices, in order to create cellular patterns for stem cell differentiation and tissue engineering.
Finally magnetic nanoparticles show also great promises for antitumor cell therapies, in particular using the magnetic hyperthermia modality. Cellular internalization of magnetic nanoparticles localizes the source of heat in the internal volume of the cell, with direct application for tumor cell therapies. The combination of cell-derived vesicles with magnetic nanoparticles creates multifunctional bio-inspired nanovectors with promising potential for diagnosis and therapy.
Cinétique de traduction de ribosomes individuels par microscope de fluorescence
Karen Perronet (Laboratoire Charles Fabry, Institut d'Optique)
Le ribosome est le moteur moléculaire traduisant le code génétique de l’ARNm en protéine. La dynamique de ce processus asynchrone primordial est encore mal connue, surtout chez les eucaryotes. Pouvoir suivre des ribosomes individuels pendant la traduction est donc un enjeu important.
Nous présentons dans ce séminaire différentes stratégies permettant d’étudier la vitesse d’élongation de ribosomes individuels par microscopie de fluorescence en réflexion totale.
Nous avons tout d’abord utilisé des ribosomes procaryotes mutants que nous avons marqués avec un nano-cristal semi-conducteur fluorescent. Nous avons ainsi pu mesurer la vitesse globale de traduction d’une protéine.
Nous avons ensuite hybridé des oligonucléotides fluorescents sur l’ARNm, les départs des marqueurs signalant alors le passage des ribosomes à leurs emplacements. Nous avons alors utilisé des ribosomes eucaryotes, encore très peu étudiés à l’échelle de la molécule unique. Les ribosomes sont initialement immobilisés sur l’ARNm grâce à une IRES, structure secondaire spécifique permettant une initiation non canonique du ribosome. Puis suite à l’injection d’un système de traduction in-vitro, ils peuvent traduire la protéine et atteindre les oligonucléotides marqués, qu’ils détachent grâce à leur activité hélicase. Ces dissociations se traduisent par une disparition du signal de fluorescence des marqueurs qui diffusent hors de l’onde évanescente. Nous avons ainsi pu observer la distribution des durées de traduction jusqu’à des endroits spécifiques de l’ARNm. Nous avons pu extraire deux durées caractéristiques pour l’élongation. La première (typ. 40 s) est associée au premier cycle d’élongation, ralentie par le fait que le ribosome doit s’extraire de l’IRES. La seconde (~1s) correspond à un cycle d’élongation normal.
Polymerization of actin branched networks controls the organization of WASH domains at the surface of endosomes
Emmanuèle Helfer (Laboratoire d'Enzymologie et de Biochimie Structurales - Gif-sur-Yvette)
Sorting of cargoes in endosomes occurs through their selective enrichment into sorting platforms, where transport intermediates are generated. The WASH complex, which is recruited from the cytosol to endosomes, activates the Arp2/3 complex and hence actin polymerization onto such sorting platforms.
Here, we analyzed the role of actin polymerization in the physiology of endosomal domains containing WASH using quantitative image analysis. Using a novel colocalization method that is insensitive to the heterogeneity of size and shape of endosomes, we show that preventing the generation of branched actin networks induces endosomal accumulation of the WASH complex. Moreover, we found that actin depolymerization induces a dramatic decrease in the recovery of endosomal WASH after photobleaching. These results suggest a built-in turnover, where the actin network, i.e. the product of the WASH complex, contributes to the dynamic cytosol/endosome exchange of the WASH complex. Our data also suggest a role of actin in the lateral compartmentalization of endosomes: discrete WASH domains coalesce upon actin depolymerization or Arp2/3 depletion. Thus, branched actin networks are involved in the regulation of WASH domain size.
I will finally discuss the potential role of lipid repartitioning in these sorting platforms and of the ensuing line tension that could develop at the domain boundary. This may provide a dynamin-independent contribution to membrane scission.
Localization and condensation of the parB-DNA complex in the bacterial chromosome
Chase Broedersz (Princeton University)
The molecular machinery responsible for faithful segregation of the chromosome in bacteria such as Caulobacter crescentus and Bacillus subtilis includes the ParABS a.k.a. Spo0J/Soj partitioning system. In Caulobacter, prior to division, hundreds of ParB proteins bind to the chromosome near the origin of replication, and localize to one pole of the cell. Subsequently, the ParB-DNA complex is translocated to the far pole by the binding and retraction of the ParA spindle-like apparatus. Remarkably, the localization of ParB proteins to specific regions of the chromosome appears to be controlled by only a few centromeric parS binding sites. Although lateral interactions between DNA-bound ParB are likely to be important for their localization, the long-range order of ParB domains on the chromosome appears to be inconsistent with a picture in which protein-protein interactions are limited to neighboring DNA-bound proteins. We propose a simple model to study the localization and condensation of the ParB-DNA complex, which includes both lateral and 3D protein-protein interactions among DNA-bound ParB proteins. Our model shows how such interactions can condense and organize the chromosome spatially, and can control both the localization and the long-range order of the DNA-bound proteins. Interestingly, the condensed ParB-DNA complex adopts a topology in which loops of DNA extend outside the cluster exposed for transcription; this structure may have implications for expression levels of genes in the vicinity of the parS site.
MicroRNAs: a selective channel of communication between competing RNAs
Andrea De Martino (Sapienza Universita' di Roma)
It has recently been suggested that the competition for a finite pool of microRNAs (miRNA) gives rise to effective interactions among their common targets (competing endogenous RNAs or ceRNAs) that could prove to be crucial for post-transcriptional regulation (PTR). I shall discuss a minimal model of PTR where the emergence and the nature of such interactions can be characterized in detail at steady state. Sensitivity analysis shows that binding free energies and repression mechanisms are the key ingredients for the cross-talk between ceRNAs to arise. Interactions emerge in specific ranges of repression values, can be symmetrical (one ceRNA influences another and vice-versa) or asymmetrical (one ceRNA influences another but not the reverse) and may be highly selective, while possibly limited by noise. Finally, I will show that non-trivial correlations among ceRNAs can emerge in experimental readouts due to transcriptional fluctuations even in absence of miRNA-mediated cross-talk, so that more refined analyses will be needed to identify signatures of cross-talk in the transcriptome.
In vitro & in vivo single molecule approaches to DNA replication
Jean-François Allemand (École normale supérieure)
DNA replication is an essential step of the cell cycle. Despite many biochemical studies, physical approaches can still bring new informations on some process of DNA replication.
In vitro, using micromanipulation tools, we have shown that force can trigger some DNA polymerases into an exonuclease mode where, instead of synthesizing DNA, the polymerase goes backward and removes nucleotides. This can lead to a mechanical method for DNA sequencing.
In vivo, in E. coli, using single molecule fluorescence techniques we have quantified the real time composition of the replisome, the machinary that copies DNA. We observed a time variation of the composition of the replisome with a number of DNA polymerases that varies from 2 to 3. We will see how these fluctuations are related to the synthesis of the so called Okazaki fragments. We will describe what happens to this scenario when the replication is blocked under specific conditions.
Nanoscopy with focused light
Stefan Hell (Max Planck Institute for Biophysical Chemistry & German Cancer Research Center (DKFZ))
SPECIAL DATE & TIME
Joint seminar with Laboratoire Aimé Cotton hosted by François Treussart
Encoding functional information into the microtubule cytoskeleton
Carsten Janke Institut Curie - Orsay
Our team is interested in the functions of tubulin posttranslational modifications. Tubulin, the building block of the microtubules, is modified by a range of very special posttranslational modifications, such as enzymatic generation of glutamate or glycine side chains, enzymatic removal and re-addition of a C-terminal tyrosine. These modifications take place once the tubulin has polymerized into microtubules, which suggests that they encode information important for the functional specification or the fine-tuning of specific microtubule functions.
While the modifications have been known for a long time, functional studies have remained difficult because nothing was known about the enzymes responsible for these modifications. The recent discovery of most of these enzymes in our team has allowed us to start functional and mechanistic studies that gave us first insights into the divergent roles of tubulin posttranslational modifications. In my presentation, I will give an overview of our first results, our ongoing projects and interests.
Properties of the actin cortex and dynamics of cytokinesis
Jean-François Joanny (Institut Curie - Paris)
In this talk I discuss first the properties of the actin cortical layer in contact with the plasma membrane of a cell using the hydrodynamic theory of active gels: within this framework, the cortical layer can be viewed as an active prewetting layer.
I then discuss cytokinesis which is last stage of cell division when the two daughter cells separate. Cytokinesis proceeds by the formation of an actin contractile ring at the equator of teh cell and by contraction of this ring. The ring formation induces tension gradients in the cortical layer and a cortical flow. Using active gel theory, it is shown numerically that cytokinesis can only be completed if the activity gradient is large enough and that the radius of the contractile ring decreases linearly with time in agreement with experiments.
Fluctuations and response in biological sensory systems
Massimo Vergassola (Institut Pasteur)
The statistics of fluctuations in biological pathways and its relation to the response to environmental stimuli will be discussed. We shall specifically focus on bacterial chemotaxis, where detailed experiments and reliable models are available. A novel non-invasive experimental method to measure the chemotaxis response will first be presented. By using fluctuation relations, we then relate appropriate steady-state correlations to the response of the system to step and ramp stimuli of arbitrary amplitudes. That provides a systematic explanation for the observed relation between fluctuations and response and it also reconciles the out-of-equilibrium nature of the dynamics with the apparently standard form of the fluctuation-dissipation relation.
Nonlinear microscopy of developing tissues and embryos
Emmanuel Beaurepaire (Lab. for Optics and Biosciences, École Polytechnique/CNRS/INSERM)
Understanding the dynamics at play during embryonic and tissue development requires tissue-scale measurements with subcellular resolution. Nonlinear microscopy is attractive for live studies because it provides tissue 3D imaging and provides several contrast mechanisms. Intrinsic coherent signals such as second- and third-harmonic generation (SHG, THG) provide structural information and can be used in conjunction with fluorescence (2PEF) imaging. SHG signals are specifically obtained from dense organized structures, whereas THG signals are obtained from optical heterogeneities and provide images of the tissue morphology. We will discuss these principles and applications such as imaging the microstructure of intact unstained corneas, and reconstructing the early development of zebrafish embryos. Finally, we will discuss ongoing efforts to improve the capabilities of nonlinear microscopy for imaging developing tissues at advanced stages, such as wavefront control for aberration correction, and simultaneous excitation of multiple fluorescent proteins.
- References:
- Supatto et al, Curr. Opin. Gen. Dev. 21 538 (2011).
- Aptel et al, Invest. Ophtalmol. Vis. Sci. 51, 2459 (2010).
- Olivier et al, Science 339, 967 (2010).
- Zeng et al, Biomed. Opt. Express 3, 1898 (2012).
- Mahou et al, Nat. Methods 9 815 (2012).
Non-equilibrium physics inspired by biology: from fluctuating membranes to active gels
Nir Gov, (Weizmann Institute of Science)
The most ubiquitous and accessible non-equilibrium systems around us are in the form of living organisms. Motivated by pattern formation and random motion that are driven by active (out-of-equilibrium) processes in cells we investigate the similarities and differences between such motion and thermal random motion. A simple theoretical model of a kicked particle is used to extract analytic and numerical results which we then compare to observations of membrane fluctuations in cells, and fluctuations in active gels reconstituted in-vitro.